
Diaminobutyricimonas aerilata
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Diaminobutyricimonas
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
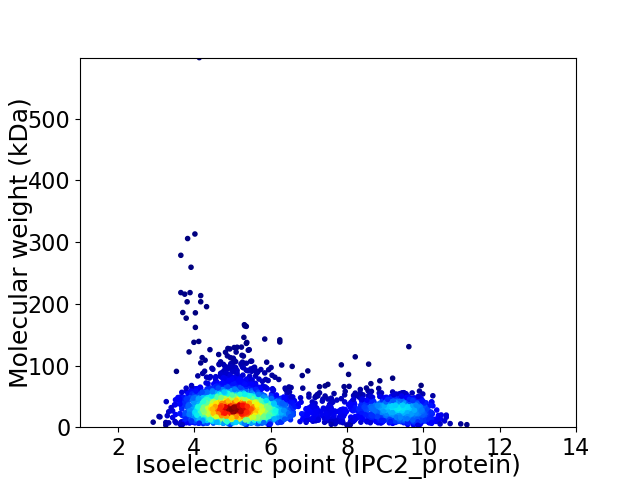
Virtual 2D-PAGE plot for 3263 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9CN69|A0A2M9CN69_9MICO Putative dehydrogenase OS=Diaminobutyricimonas aerilata OX=1162967 GN=CLV46_2943 PE=4 SV=1
MM1 pKa = 7.46GLAIATVIATAALTLAGGLPAQAVGPAEE29 pKa = 4.29LSIEE33 pKa = 4.02ITPVNPTTGEE43 pKa = 4.32VITEE47 pKa = 4.1TAFNQHH53 pKa = 5.87NDD55 pKa = 3.0RR56 pKa = 11.84LAYY59 pKa = 10.11RR60 pKa = 11.84IGYY63 pKa = 7.74SCSVAEE69 pKa = 4.38CTDD72 pKa = 3.43AVVTLPAVATDD83 pKa = 3.41PTYY86 pKa = 10.86GQFRR90 pKa = 11.84LHH92 pKa = 7.19HH93 pKa = 6.25YY94 pKa = 7.33EE95 pKa = 3.65TWTPPAGGGATITNSEE111 pKa = 4.22TAGITVNLGTLPAGTSATFQVQYY134 pKa = 10.82IRR136 pKa = 11.84YY137 pKa = 7.45TGGSAPNPLEE147 pKa = 3.92AAYY150 pKa = 9.35FPPGYY155 pKa = 9.92QIEE158 pKa = 4.27RR159 pKa = 11.84SATISSPNATADD171 pKa = 3.37ATAVASPVTWNIQIPTPAIGKK192 pKa = 9.56VGLTATTRR200 pKa = 11.84PDD202 pKa = 2.63TDD204 pKa = 3.33YY205 pKa = 10.45TYY207 pKa = 10.67RR208 pKa = 11.84IAMSDD213 pKa = 3.44GCFVNQGAARR223 pKa = 11.84WLATGPYY230 pKa = 9.65LCAEE234 pKa = 4.35NFTVVDD240 pKa = 4.0TLPDD244 pKa = 3.23QAEE247 pKa = 4.32FVSATGGGVYY257 pKa = 10.3DD258 pKa = 4.69PEE260 pKa = 4.5THH262 pKa = 5.44TVTWTGSGPRR272 pKa = 11.84AAGGWGVALYY282 pKa = 9.14TGWSTGNGYY291 pKa = 11.07ANRR294 pKa = 11.84FVTVRR299 pKa = 11.84FPADD303 pKa = 3.7AFPEE307 pKa = 4.37AAGGADD313 pKa = 4.79FIAPVTNDD321 pKa = 2.53IEE323 pKa = 4.4ATVTYY328 pKa = 10.53LDD330 pKa = 4.59DD331 pKa = 4.23AQTTKK336 pKa = 10.36TVTASSEE343 pKa = 4.13NQVIRR348 pKa = 11.84AAPFGRR354 pKa = 11.84ANQTKK359 pKa = 8.63DD360 pKa = 3.11TTSDD364 pKa = 3.41QIIGGQRR371 pKa = 11.84FVNVPPDD378 pKa = 3.65TTGLVCPEE386 pKa = 4.59SGRR389 pKa = 11.84DD390 pKa = 3.46DD391 pKa = 3.36WGTTCTPGGAVATYY405 pKa = 7.53PTQLEE410 pKa = 4.34YY411 pKa = 10.44WQVDD415 pKa = 3.66TYY417 pKa = 11.91NRR419 pKa = 11.84GNVPGVAVITDD430 pKa = 3.83DD431 pKa = 5.39DD432 pKa = 5.1LGDD435 pKa = 3.68SALRR439 pKa = 11.84VYY441 pKa = 10.42QINTSAAATIAVTLSDD457 pKa = 3.95GTPARR462 pKa = 11.84ATGTQYY468 pKa = 10.97VAPAGLRR475 pKa = 11.84IIAATVTSGPIAGPNLLPSQNAGTLFRR502 pKa = 11.84TLYY505 pKa = 10.04RR506 pKa = 11.84YY507 pKa = 7.34EE508 pKa = 4.29VPVGAPTDD516 pKa = 3.88VAWTNTATATMSYY529 pKa = 10.52PEE531 pKa = 4.26YY532 pKa = 10.41PEE534 pKa = 4.42IADD537 pKa = 3.43IPMEE541 pKa = 4.3ATATANFRR549 pKa = 11.84DD550 pKa = 4.3WPTVTVPVTPPTFGAGFASAPVVEE574 pKa = 5.19GGGQVVPGGRR584 pKa = 11.84VTFGVRR590 pKa = 11.84GGTANIPADD599 pKa = 3.95RR600 pKa = 11.84DD601 pKa = 3.5VSPQYY606 pKa = 11.64VFIAPVGWAVVPNSASFPAGSVPPGVAYY634 pKa = 9.12TYY636 pKa = 11.17KK637 pKa = 10.5DD638 pKa = 3.3VTIAGVTRR646 pKa = 11.84QAVIASWPTGTTFGKK661 pKa = 10.18NVTWPTMNVVASPTSAVAAGTNSVATAWAGDD692 pKa = 3.48SRR694 pKa = 11.84NAYY697 pKa = 9.85EE698 pKa = 5.39PDD700 pKa = 3.21TTTWNVKK707 pKa = 9.79VVDD710 pKa = 3.98TTDD713 pKa = 2.77VDD715 pKa = 3.7GDD717 pKa = 4.23GNSTEE722 pKa = 4.86AFAAGNSAAVLVSGTSRR739 pKa = 11.84LDD741 pKa = 3.45TVKK744 pKa = 10.35EE745 pKa = 3.84ICVLDD750 pKa = 3.78EE751 pKa = 5.55DD752 pKa = 5.64GACEE756 pKa = 4.05WVSNPDD762 pKa = 2.59IVAGVDD768 pKa = 3.41PDD770 pKa = 3.78ATDD773 pKa = 2.76ITYY776 pKa = 10.48RR777 pKa = 11.84ITFSNGSNQTLSNVVGYY794 pKa = 10.5DD795 pKa = 3.18VLPYY799 pKa = 10.76VGDD802 pKa = 3.7PRR804 pKa = 11.84GSTFGEE810 pKa = 4.24TLNEE814 pKa = 4.19VTSVSPNLEE823 pKa = 3.85LSYY826 pKa = 10.9SASTNPCRR834 pKa = 11.84PEE836 pKa = 3.89VLSTNPGCEE845 pKa = 4.15TGWTGSADD853 pKa = 3.43GARR856 pKa = 11.84SIRR859 pKa = 11.84AALTGTLAPGASASFTFTASVVPGAAADD887 pKa = 3.81AVACNSVASDD897 pKa = 3.65SASTLPAEE905 pKa = 4.52PRR907 pKa = 11.84PVCATTQEE915 pKa = 3.78ADD917 pKa = 3.67LAVTVPSRR925 pKa = 11.84LPLQAEE931 pKa = 4.29RR932 pKa = 11.84PGVVPFTVANLGPSANAPATVEE954 pKa = 3.69IDD956 pKa = 3.46VPAGIRR962 pKa = 11.84ITSLDD967 pKa = 3.49PDD969 pKa = 3.86GWVCAADD976 pKa = 3.73EE977 pKa = 4.37VEE979 pKa = 4.49PDD981 pKa = 3.62GSVLGPVTLSCEE993 pKa = 3.96PVTAEE998 pKa = 3.59GTARR1002 pKa = 11.84MLAIDD1007 pKa = 4.67EE1008 pKa = 4.73PNALDD1013 pKa = 4.14LPAVIPSDD1021 pKa = 3.56EE1022 pKa = 4.18LVGEE1026 pKa = 4.76GACFAAAVSGLMSDD1040 pKa = 3.63PVEE1043 pKa = 4.45EE1044 pKa = 4.33NDD1046 pKa = 3.55TAEE1049 pKa = 3.92ACFDD1053 pKa = 3.84VVQGDD1058 pKa = 4.16TLVGLTKK1065 pKa = 10.59DD1066 pKa = 3.79DD1067 pKa = 4.03GLGSVAMGEE1076 pKa = 4.24EE1077 pKa = 4.1FTYY1080 pKa = 10.44TIDD1083 pKa = 3.79VANLLVGEE1091 pKa = 4.36GLSDD1095 pKa = 3.88IVITDD1100 pKa = 3.83EE1101 pKa = 5.43LPPTLAFVSASAGGSIEE1118 pKa = 4.12GQGDD1122 pKa = 3.5ADD1124 pKa = 4.35ADD1126 pKa = 3.83GLRR1129 pKa = 11.84AGGTVTWNLASLAASGQPDD1148 pKa = 3.69ADD1150 pKa = 3.48GDD1152 pKa = 4.2VAAGAAGSTQQLTVTVRR1169 pKa = 11.84VVQAAEE1175 pKa = 3.97AVGEE1179 pKa = 4.21IANTATVTTIDD1190 pKa = 3.52PALPEE1195 pKa = 4.29VVLTATDD1202 pKa = 3.92DD1203 pKa = 4.43DD1204 pKa = 4.31VDD1206 pKa = 3.88EE1207 pKa = 5.56LIRR1210 pKa = 11.84TAGLTLVKK1218 pKa = 9.56TADD1221 pKa = 3.75PVTVDD1226 pKa = 3.3SVGDD1230 pKa = 3.63VITYY1234 pKa = 10.62SFLATNSGDD1243 pKa = 3.6VTLSAVGVTEE1253 pKa = 4.02TAFTGTGIAPAVTCPSEE1270 pKa = 4.14AATLAPAASVTCTATYY1286 pKa = 9.62TVTQADD1292 pKa = 4.19LDD1294 pKa = 4.13AGIVTNTATAAATAPAGVEE1313 pKa = 4.34GPVSNASTATVTAQALPALTLVKK1336 pKa = 10.45SAGDD1340 pKa = 3.65AVIDD1344 pKa = 3.96SAGDD1348 pKa = 3.47TVSYY1352 pKa = 10.8LFLVTNTGNVTLSDD1366 pKa = 3.64VAIDD1370 pKa = 3.45EE1371 pKa = 4.52TAFTGTGADD1380 pKa = 3.56VVAEE1384 pKa = 4.18CADD1387 pKa = 3.57GVLAPGASVICTASYY1402 pKa = 10.88EE1403 pKa = 4.35VTQPDD1408 pKa = 3.87VNAGRR1413 pKa = 11.84IDD1415 pKa = 3.63NTAVAVATTPTDD1427 pKa = 3.35APVSSEE1433 pKa = 3.91ASSASLVIGAAPALSVVKK1451 pKa = 10.51SATPTDD1457 pKa = 3.66LVVDD1461 pKa = 3.71QEE1463 pKa = 4.42VTYY1466 pKa = 11.16SFVVTNTGNVTLDD1479 pKa = 3.38DD1480 pKa = 3.87VAIDD1484 pKa = 3.6EE1485 pKa = 4.79VEE1487 pKa = 4.29FTGSGALSDD1496 pKa = 4.1VVCAAGAGALDD1507 pKa = 4.88PGDD1510 pKa = 3.9QVICSATYY1518 pKa = 9.44TITQADD1524 pKa = 3.62VDD1526 pKa = 4.83AGTLSNTATATAVAPGGPLTSEE1548 pKa = 4.19PSSVQLPFVQEE1559 pKa = 4.04PGVSVVKK1566 pKa = 10.7SADD1569 pKa = 3.16VDD1571 pKa = 4.23GYY1573 pKa = 11.34AAAGDD1578 pKa = 3.55AVEE1581 pKa = 3.94YY1582 pKa = 10.29RR1583 pKa = 11.84FRR1585 pKa = 11.84VTNTGNVSLADD1596 pKa = 3.66AEE1598 pKa = 4.91VIEE1601 pKa = 4.51QDD1603 pKa = 3.87FSGTGDD1609 pKa = 4.14LSAVDD1614 pKa = 4.3CATGSLLPGQFVDD1627 pKa = 4.12CTADD1631 pKa = 3.66YY1632 pKa = 10.73EE1633 pKa = 4.66VTQADD1638 pKa = 3.43VDD1640 pKa = 3.95AGEE1643 pKa = 4.56LTNSATATGVAPGSADD1659 pKa = 3.83PIEE1662 pKa = 5.03SEE1664 pKa = 4.33LSEE1667 pKa = 4.36LVLPFTGEE1675 pKa = 3.78LALTLTKK1682 pKa = 10.63AGTPVDD1688 pKa = 3.32VDD1690 pKa = 3.27GDD1692 pKa = 3.97GRR1694 pKa = 11.84TTAADD1699 pKa = 4.84RR1700 pKa = 11.84IQWSFTVTNPSAATLSDD1717 pKa = 3.58VTVVDD1722 pKa = 4.44PMAGEE1727 pKa = 4.49IVCDD1731 pKa = 3.82ATVLAPGDD1739 pKa = 4.26SVDD1742 pKa = 3.63CAAVEE1747 pKa = 4.66EE1748 pKa = 4.32YY1749 pKa = 10.78QITAQQAAAGQVLNVASAFAVGVGNVEE1776 pKa = 3.93VAAAEE1781 pKa = 4.05AEE1783 pKa = 4.23AVVEE1787 pKa = 4.47VVTVPPAAAGPGTGGLVSTGVEE1809 pKa = 3.93VRR1811 pKa = 11.84LTLLLGLLALLAGIGLYY1828 pKa = 9.5VAQRR1832 pKa = 11.84RR1833 pKa = 11.84RR1834 pKa = 11.84TRR1836 pKa = 11.84QAA1838 pKa = 2.64
MM1 pKa = 7.46GLAIATVIATAALTLAGGLPAQAVGPAEE29 pKa = 4.29LSIEE33 pKa = 4.02ITPVNPTTGEE43 pKa = 4.32VITEE47 pKa = 4.1TAFNQHH53 pKa = 5.87NDD55 pKa = 3.0RR56 pKa = 11.84LAYY59 pKa = 10.11RR60 pKa = 11.84IGYY63 pKa = 7.74SCSVAEE69 pKa = 4.38CTDD72 pKa = 3.43AVVTLPAVATDD83 pKa = 3.41PTYY86 pKa = 10.86GQFRR90 pKa = 11.84LHH92 pKa = 7.19HH93 pKa = 6.25YY94 pKa = 7.33EE95 pKa = 3.65TWTPPAGGGATITNSEE111 pKa = 4.22TAGITVNLGTLPAGTSATFQVQYY134 pKa = 10.82IRR136 pKa = 11.84YY137 pKa = 7.45TGGSAPNPLEE147 pKa = 3.92AAYY150 pKa = 9.35FPPGYY155 pKa = 9.92QIEE158 pKa = 4.27RR159 pKa = 11.84SATISSPNATADD171 pKa = 3.37ATAVASPVTWNIQIPTPAIGKK192 pKa = 9.56VGLTATTRR200 pKa = 11.84PDD202 pKa = 2.63TDD204 pKa = 3.33YY205 pKa = 10.45TYY207 pKa = 10.67RR208 pKa = 11.84IAMSDD213 pKa = 3.44GCFVNQGAARR223 pKa = 11.84WLATGPYY230 pKa = 9.65LCAEE234 pKa = 4.35NFTVVDD240 pKa = 4.0TLPDD244 pKa = 3.23QAEE247 pKa = 4.32FVSATGGGVYY257 pKa = 10.3DD258 pKa = 4.69PEE260 pKa = 4.5THH262 pKa = 5.44TVTWTGSGPRR272 pKa = 11.84AAGGWGVALYY282 pKa = 9.14TGWSTGNGYY291 pKa = 11.07ANRR294 pKa = 11.84FVTVRR299 pKa = 11.84FPADD303 pKa = 3.7AFPEE307 pKa = 4.37AAGGADD313 pKa = 4.79FIAPVTNDD321 pKa = 2.53IEE323 pKa = 4.4ATVTYY328 pKa = 10.53LDD330 pKa = 4.59DD331 pKa = 4.23AQTTKK336 pKa = 10.36TVTASSEE343 pKa = 4.13NQVIRR348 pKa = 11.84AAPFGRR354 pKa = 11.84ANQTKK359 pKa = 8.63DD360 pKa = 3.11TTSDD364 pKa = 3.41QIIGGQRR371 pKa = 11.84FVNVPPDD378 pKa = 3.65TTGLVCPEE386 pKa = 4.59SGRR389 pKa = 11.84DD390 pKa = 3.46DD391 pKa = 3.36WGTTCTPGGAVATYY405 pKa = 7.53PTQLEE410 pKa = 4.34YY411 pKa = 10.44WQVDD415 pKa = 3.66TYY417 pKa = 11.91NRR419 pKa = 11.84GNVPGVAVITDD430 pKa = 3.83DD431 pKa = 5.39DD432 pKa = 5.1LGDD435 pKa = 3.68SALRR439 pKa = 11.84VYY441 pKa = 10.42QINTSAAATIAVTLSDD457 pKa = 3.95GTPARR462 pKa = 11.84ATGTQYY468 pKa = 10.97VAPAGLRR475 pKa = 11.84IIAATVTSGPIAGPNLLPSQNAGTLFRR502 pKa = 11.84TLYY505 pKa = 10.04RR506 pKa = 11.84YY507 pKa = 7.34EE508 pKa = 4.29VPVGAPTDD516 pKa = 3.88VAWTNTATATMSYY529 pKa = 10.52PEE531 pKa = 4.26YY532 pKa = 10.41PEE534 pKa = 4.42IADD537 pKa = 3.43IPMEE541 pKa = 4.3ATATANFRR549 pKa = 11.84DD550 pKa = 4.3WPTVTVPVTPPTFGAGFASAPVVEE574 pKa = 5.19GGGQVVPGGRR584 pKa = 11.84VTFGVRR590 pKa = 11.84GGTANIPADD599 pKa = 3.95RR600 pKa = 11.84DD601 pKa = 3.5VSPQYY606 pKa = 11.64VFIAPVGWAVVPNSASFPAGSVPPGVAYY634 pKa = 9.12TYY636 pKa = 11.17KK637 pKa = 10.5DD638 pKa = 3.3VTIAGVTRR646 pKa = 11.84QAVIASWPTGTTFGKK661 pKa = 10.18NVTWPTMNVVASPTSAVAAGTNSVATAWAGDD692 pKa = 3.48SRR694 pKa = 11.84NAYY697 pKa = 9.85EE698 pKa = 5.39PDD700 pKa = 3.21TTTWNVKK707 pKa = 9.79VVDD710 pKa = 3.98TTDD713 pKa = 2.77VDD715 pKa = 3.7GDD717 pKa = 4.23GNSTEE722 pKa = 4.86AFAAGNSAAVLVSGTSRR739 pKa = 11.84LDD741 pKa = 3.45TVKK744 pKa = 10.35EE745 pKa = 3.84ICVLDD750 pKa = 3.78EE751 pKa = 5.55DD752 pKa = 5.64GACEE756 pKa = 4.05WVSNPDD762 pKa = 2.59IVAGVDD768 pKa = 3.41PDD770 pKa = 3.78ATDD773 pKa = 2.76ITYY776 pKa = 10.48RR777 pKa = 11.84ITFSNGSNQTLSNVVGYY794 pKa = 10.5DD795 pKa = 3.18VLPYY799 pKa = 10.76VGDD802 pKa = 3.7PRR804 pKa = 11.84GSTFGEE810 pKa = 4.24TLNEE814 pKa = 4.19VTSVSPNLEE823 pKa = 3.85LSYY826 pKa = 10.9SASTNPCRR834 pKa = 11.84PEE836 pKa = 3.89VLSTNPGCEE845 pKa = 4.15TGWTGSADD853 pKa = 3.43GARR856 pKa = 11.84SIRR859 pKa = 11.84AALTGTLAPGASASFTFTASVVPGAAADD887 pKa = 3.81AVACNSVASDD897 pKa = 3.65SASTLPAEE905 pKa = 4.52PRR907 pKa = 11.84PVCATTQEE915 pKa = 3.78ADD917 pKa = 3.67LAVTVPSRR925 pKa = 11.84LPLQAEE931 pKa = 4.29RR932 pKa = 11.84PGVVPFTVANLGPSANAPATVEE954 pKa = 3.69IDD956 pKa = 3.46VPAGIRR962 pKa = 11.84ITSLDD967 pKa = 3.49PDD969 pKa = 3.86GWVCAADD976 pKa = 3.73EE977 pKa = 4.37VEE979 pKa = 4.49PDD981 pKa = 3.62GSVLGPVTLSCEE993 pKa = 3.96PVTAEE998 pKa = 3.59GTARR1002 pKa = 11.84MLAIDD1007 pKa = 4.67EE1008 pKa = 4.73PNALDD1013 pKa = 4.14LPAVIPSDD1021 pKa = 3.56EE1022 pKa = 4.18LVGEE1026 pKa = 4.76GACFAAAVSGLMSDD1040 pKa = 3.63PVEE1043 pKa = 4.45EE1044 pKa = 4.33NDD1046 pKa = 3.55TAEE1049 pKa = 3.92ACFDD1053 pKa = 3.84VVQGDD1058 pKa = 4.16TLVGLTKK1065 pKa = 10.59DD1066 pKa = 3.79DD1067 pKa = 4.03GLGSVAMGEE1076 pKa = 4.24EE1077 pKa = 4.1FTYY1080 pKa = 10.44TIDD1083 pKa = 3.79VANLLVGEE1091 pKa = 4.36GLSDD1095 pKa = 3.88IVITDD1100 pKa = 3.83EE1101 pKa = 5.43LPPTLAFVSASAGGSIEE1118 pKa = 4.12GQGDD1122 pKa = 3.5ADD1124 pKa = 4.35ADD1126 pKa = 3.83GLRR1129 pKa = 11.84AGGTVTWNLASLAASGQPDD1148 pKa = 3.69ADD1150 pKa = 3.48GDD1152 pKa = 4.2VAAGAAGSTQQLTVTVRR1169 pKa = 11.84VVQAAEE1175 pKa = 3.97AVGEE1179 pKa = 4.21IANTATVTTIDD1190 pKa = 3.52PALPEE1195 pKa = 4.29VVLTATDD1202 pKa = 3.92DD1203 pKa = 4.43DD1204 pKa = 4.31VDD1206 pKa = 3.88EE1207 pKa = 5.56LIRR1210 pKa = 11.84TAGLTLVKK1218 pKa = 9.56TADD1221 pKa = 3.75PVTVDD1226 pKa = 3.3SVGDD1230 pKa = 3.63VITYY1234 pKa = 10.62SFLATNSGDD1243 pKa = 3.6VTLSAVGVTEE1253 pKa = 4.02TAFTGTGIAPAVTCPSEE1270 pKa = 4.14AATLAPAASVTCTATYY1286 pKa = 9.62TVTQADD1292 pKa = 4.19LDD1294 pKa = 4.13AGIVTNTATAAATAPAGVEE1313 pKa = 4.34GPVSNASTATVTAQALPALTLVKK1336 pKa = 10.45SAGDD1340 pKa = 3.65AVIDD1344 pKa = 3.96SAGDD1348 pKa = 3.47TVSYY1352 pKa = 10.8LFLVTNTGNVTLSDD1366 pKa = 3.64VAIDD1370 pKa = 3.45EE1371 pKa = 4.52TAFTGTGADD1380 pKa = 3.56VVAEE1384 pKa = 4.18CADD1387 pKa = 3.57GVLAPGASVICTASYY1402 pKa = 10.88EE1403 pKa = 4.35VTQPDD1408 pKa = 3.87VNAGRR1413 pKa = 11.84IDD1415 pKa = 3.63NTAVAVATTPTDD1427 pKa = 3.35APVSSEE1433 pKa = 3.91ASSASLVIGAAPALSVVKK1451 pKa = 10.51SATPTDD1457 pKa = 3.66LVVDD1461 pKa = 3.71QEE1463 pKa = 4.42VTYY1466 pKa = 11.16SFVVTNTGNVTLDD1479 pKa = 3.38DD1480 pKa = 3.87VAIDD1484 pKa = 3.6EE1485 pKa = 4.79VEE1487 pKa = 4.29FTGSGALSDD1496 pKa = 4.1VVCAAGAGALDD1507 pKa = 4.88PGDD1510 pKa = 3.9QVICSATYY1518 pKa = 9.44TITQADD1524 pKa = 3.62VDD1526 pKa = 4.83AGTLSNTATATAVAPGGPLTSEE1548 pKa = 4.19PSSVQLPFVQEE1559 pKa = 4.04PGVSVVKK1566 pKa = 10.7SADD1569 pKa = 3.16VDD1571 pKa = 4.23GYY1573 pKa = 11.34AAAGDD1578 pKa = 3.55AVEE1581 pKa = 3.94YY1582 pKa = 10.29RR1583 pKa = 11.84FRR1585 pKa = 11.84VTNTGNVSLADD1596 pKa = 3.66AEE1598 pKa = 4.91VIEE1601 pKa = 4.51QDD1603 pKa = 3.87FSGTGDD1609 pKa = 4.14LSAVDD1614 pKa = 4.3CATGSLLPGQFVDD1627 pKa = 4.12CTADD1631 pKa = 3.66YY1632 pKa = 10.73EE1633 pKa = 4.66VTQADD1638 pKa = 3.43VDD1640 pKa = 3.95AGEE1643 pKa = 4.56LTNSATATGVAPGSADD1659 pKa = 3.83PIEE1662 pKa = 5.03SEE1664 pKa = 4.33LSEE1667 pKa = 4.36LVLPFTGEE1675 pKa = 3.78LALTLTKK1682 pKa = 10.63AGTPVDD1688 pKa = 3.32VDD1690 pKa = 3.27GDD1692 pKa = 3.97GRR1694 pKa = 11.84TTAADD1699 pKa = 4.84RR1700 pKa = 11.84IQWSFTVTNPSAATLSDD1717 pKa = 3.58VTVVDD1722 pKa = 4.44PMAGEE1727 pKa = 4.49IVCDD1731 pKa = 3.82ATVLAPGDD1739 pKa = 4.26SVDD1742 pKa = 3.63CAAVEE1747 pKa = 4.66EE1748 pKa = 4.32YY1749 pKa = 10.78QITAQQAAAGQVLNVASAFAVGVGNVEE1776 pKa = 3.93VAAAEE1781 pKa = 4.05AEE1783 pKa = 4.23AVVEE1787 pKa = 4.47VVTVPPAAAGPGTGGLVSTGVEE1809 pKa = 3.93VRR1811 pKa = 11.84LTLLLGLLALLAGIGLYY1828 pKa = 9.5VAQRR1832 pKa = 11.84RR1833 pKa = 11.84RR1834 pKa = 11.84TRR1836 pKa = 11.84QAA1838 pKa = 2.64
Molecular weight: 185.92 kDa
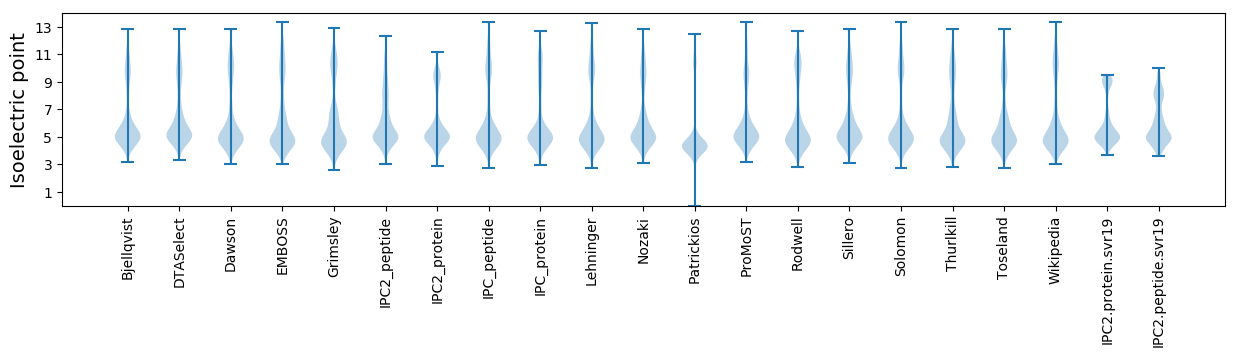
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9CGA0|A0A2M9CGA0_9MICO Uncharacterized protein OS=Diaminobutyricimonas aerilata OX=1162967 GN=CLV46_0419 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1073911 |
29 |
6075 |
329.1 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.386 ± 0.063 | 0.436 ± 0.01 |
6.385 ± 0.043 | 5.848 ± 0.05 |
3.123 ± 0.029 | 8.96 ± 0.042 |
1.941 ± 0.024 | 4.214 ± 0.033 |
1.64 ± 0.031 | 10.237 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.64 ± 0.021 | 1.923 ± 0.029 |
5.568 ± 0.038 | 2.627 ± 0.024 |
7.676 ± 0.065 | 5.379 ± 0.03 |
6.201 ± 0.083 | 9.276 ± 0.046 |
1.516 ± 0.021 | 2.024 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
