
Hibiscus green spot virus (isolate Citrus volkameriana /USA/WAI 1-1/2009)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Kitaviridae; Higrevirus; unclassified Higrevirus; Hibiscus green spot virus
Average proteome isoelectric point is 7.23
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G8DPK0|G8DPK0_HGSV2 p9 OS=Hibiscus green spot virus (isolate Citrus volkameriana /USA/WAI 1-1/2009) OX=1554497 PE=4 SV=1
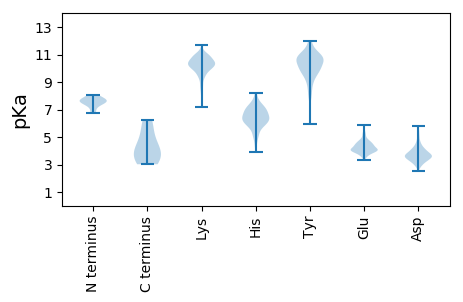
MM1 pKa = 7.72EE2 pKa = 4.75SFNYY6 pKa = 7.68VTSADD11 pKa = 4.16LVCSLSRR18 pKa = 11.84FRR20 pKa = 11.84DD21 pKa = 4.52FYY23 pKa = 11.2KK24 pKa = 10.59EE25 pKa = 3.83RR26 pKa = 11.84SASLSDD32 pKa = 3.55DD33 pKa = 3.88DD34 pKa = 5.82VYY36 pKa = 11.95ADD38 pKa = 3.51TTRR41 pKa = 11.84EE42 pKa = 3.56YY43 pKa = 11.2SAALDD48 pKa = 3.38STEE51 pKa = 3.72FVDD54 pKa = 4.67YY55 pKa = 11.04NFQIFLLNGVPGGGKK70 pKa = 8.17TKK72 pKa = 10.26FVYY75 pKa = 10.63EE76 pKa = 4.03NVEE79 pKa = 3.88VRR81 pKa = 11.84NSCVVVPFKK90 pKa = 10.85ALKK93 pKa = 10.3DD94 pKa = 3.66EE95 pKa = 4.04YY96 pKa = 10.49RR97 pKa = 11.84KK98 pKa = 10.11RR99 pKa = 11.84GYY101 pKa = 10.61RR102 pKa = 11.84SFTQFRR108 pKa = 11.84ALPSVRR114 pKa = 11.84SADD117 pKa = 3.0ILVIDD122 pKa = 5.54EE123 pKa = 4.48YY124 pKa = 11.44TCVCYY129 pKa = 10.43SVLVSLVYY137 pKa = 10.45KK138 pKa = 10.52LRR140 pKa = 11.84PATVALIGDD149 pKa = 5.15FNQCWIRR156 pKa = 11.84DD157 pKa = 3.79GEE159 pKa = 4.47GFSMEE164 pKa = 4.5PFVSTLVVNKK174 pKa = 9.77YY175 pKa = 10.67LSVCYY180 pKa = 9.82RR181 pKa = 11.84CPCPDD186 pKa = 3.01VEE188 pKa = 4.94FVRR191 pKa = 11.84NYY193 pKa = 10.8LDD195 pKa = 5.44LDD197 pKa = 3.57ITPGKK202 pKa = 10.18CGSTCSCTGFNVEE215 pKa = 4.41TLVEE219 pKa = 4.77GMTFDD224 pKa = 3.7YY225 pKa = 10.42RR226 pKa = 11.84GEE228 pKa = 4.07VALVFSGASKK238 pKa = 10.8SYY240 pKa = 10.55LEE242 pKa = 4.67SIGVSAVTVRR252 pKa = 11.84SFMGKK257 pKa = 9.23QADD260 pKa = 4.01TVALFILKK268 pKa = 10.28DD269 pKa = 3.24HH270 pKa = 7.77DD271 pKa = 3.67IRR273 pKa = 11.84LLGVSSLKK281 pKa = 10.5LVAFTRR287 pKa = 11.84HH288 pKa = 3.91VSKK291 pKa = 10.96LVVYY295 pKa = 11.01SDD297 pKa = 3.82MSHH300 pKa = 7.29DD301 pKa = 4.1DD302 pKa = 3.53VVALIEE308 pKa = 4.38GATEE312 pKa = 3.78SGEE315 pKa = 4.62VVDD318 pKa = 5.58HH319 pKa = 7.98DD320 pKa = 4.27NFQPAYY326 pKa = 9.29EE327 pKa = 5.14DD328 pKa = 3.58SLEE331 pKa = 4.11QYY333 pKa = 10.78FDD335 pKa = 3.36RR336 pKa = 11.84LLRR339 pKa = 11.84GDD341 pKa = 4.45FYY343 pKa = 11.95VLL345 pKa = 3.41
MM1 pKa = 7.72EE2 pKa = 4.75SFNYY6 pKa = 7.68VTSADD11 pKa = 4.16LVCSLSRR18 pKa = 11.84FRR20 pKa = 11.84DD21 pKa = 4.52FYY23 pKa = 11.2KK24 pKa = 10.59EE25 pKa = 3.83RR26 pKa = 11.84SASLSDD32 pKa = 3.55DD33 pKa = 3.88DD34 pKa = 5.82VYY36 pKa = 11.95ADD38 pKa = 3.51TTRR41 pKa = 11.84EE42 pKa = 3.56YY43 pKa = 11.2SAALDD48 pKa = 3.38STEE51 pKa = 3.72FVDD54 pKa = 4.67YY55 pKa = 11.04NFQIFLLNGVPGGGKK70 pKa = 8.17TKK72 pKa = 10.26FVYY75 pKa = 10.63EE76 pKa = 4.03NVEE79 pKa = 3.88VRR81 pKa = 11.84NSCVVVPFKK90 pKa = 10.85ALKK93 pKa = 10.3DD94 pKa = 3.66EE95 pKa = 4.04YY96 pKa = 10.49RR97 pKa = 11.84KK98 pKa = 10.11RR99 pKa = 11.84GYY101 pKa = 10.61RR102 pKa = 11.84SFTQFRR108 pKa = 11.84ALPSVRR114 pKa = 11.84SADD117 pKa = 3.0ILVIDD122 pKa = 5.54EE123 pKa = 4.48YY124 pKa = 11.44TCVCYY129 pKa = 10.43SVLVSLVYY137 pKa = 10.45KK138 pKa = 10.52LRR140 pKa = 11.84PATVALIGDD149 pKa = 5.15FNQCWIRR156 pKa = 11.84DD157 pKa = 3.79GEE159 pKa = 4.47GFSMEE164 pKa = 4.5PFVSTLVVNKK174 pKa = 9.77YY175 pKa = 10.67LSVCYY180 pKa = 9.82RR181 pKa = 11.84CPCPDD186 pKa = 3.01VEE188 pKa = 4.94FVRR191 pKa = 11.84NYY193 pKa = 10.8LDD195 pKa = 5.44LDD197 pKa = 3.57ITPGKK202 pKa = 10.18CGSTCSCTGFNVEE215 pKa = 4.41TLVEE219 pKa = 4.77GMTFDD224 pKa = 3.7YY225 pKa = 10.42RR226 pKa = 11.84GEE228 pKa = 4.07VALVFSGASKK238 pKa = 10.8SYY240 pKa = 10.55LEE242 pKa = 4.67SIGVSAVTVRR252 pKa = 11.84SFMGKK257 pKa = 9.23QADD260 pKa = 4.01TVALFILKK268 pKa = 10.28DD269 pKa = 3.24HH270 pKa = 7.77DD271 pKa = 3.67IRR273 pKa = 11.84LLGVSSLKK281 pKa = 10.5LVAFTRR287 pKa = 11.84HH288 pKa = 3.91VSKK291 pKa = 10.96LVVYY295 pKa = 11.01SDD297 pKa = 3.82MSHH300 pKa = 7.29DD301 pKa = 4.1DD302 pKa = 3.53VVALIEE308 pKa = 4.38GATEE312 pKa = 3.78SGEE315 pKa = 4.62VVDD318 pKa = 5.58HH319 pKa = 7.98DD320 pKa = 4.27NFQPAYY326 pKa = 9.29EE327 pKa = 5.14DD328 pKa = 3.58SLEE331 pKa = 4.11QYY333 pKa = 10.78FDD335 pKa = 3.36RR336 pKa = 11.84LLRR339 pKa = 11.84GDD341 pKa = 4.45FYY343 pKa = 11.95VLL345 pKa = 3.41
Molecular weight: 38.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8DPK4|G8DPK4_HGSV2 p23 OS=Hibiscus green spot virus (isolate Citrus volkameriana /USA/WAI 1-1/2009) OX=1554497 PE=4 SV=1
MM1 pKa = 7.59LGLPEE6 pKa = 4.04LRR8 pKa = 11.84LRR10 pKa = 11.84LRR12 pKa = 11.84ARR14 pKa = 11.84GKK16 pKa = 10.89GLDD19 pKa = 3.44ALIYY23 pKa = 10.56DD24 pKa = 4.52VFNICLKK31 pKa = 10.88FIMKK35 pKa = 8.68PHH37 pKa = 6.52ILIMYY42 pKa = 8.08ACLLVVVYY50 pKa = 10.65NKK52 pKa = 10.92GEE54 pKa = 4.16AGDD57 pKa = 4.18VIEE60 pKa = 5.21KK61 pKa = 10.4ALKK64 pKa = 9.9AYY66 pKa = 8.41PRR68 pKa = 11.84NPILKK73 pKa = 8.56WAKK76 pKa = 7.64TDD78 pKa = 3.36YY79 pKa = 11.03VRR81 pKa = 11.84FIGILVGLPVIIDD94 pKa = 3.74APRR97 pKa = 11.84SFQLLITGAVFVSTYY112 pKa = 10.31ILPPQTLSTYY122 pKa = 10.92LLGFVSLHH130 pKa = 6.17VYY132 pKa = 10.5SKK134 pKa = 11.12AKK136 pKa = 10.07SVQLRR141 pKa = 11.84IILLASFATYY151 pKa = 10.49LVLSGIVDD159 pKa = 3.71TEE161 pKa = 4.13KK162 pKa = 10.23FTGSGTVLGGAHH174 pKa = 7.3GAPKK178 pKa = 10.21LSEE181 pKa = 4.15VPIPTVLNDD190 pKa = 3.84PGPRR194 pKa = 11.84IRR196 pKa = 11.84SVEE199 pKa = 3.64EE200 pKa = 3.41RR201 pKa = 11.84LPVIKK206 pKa = 10.32RR207 pKa = 11.84GGSGGVV213 pKa = 3.02
MM1 pKa = 7.59LGLPEE6 pKa = 4.04LRR8 pKa = 11.84LRR10 pKa = 11.84LRR12 pKa = 11.84ARR14 pKa = 11.84GKK16 pKa = 10.89GLDD19 pKa = 3.44ALIYY23 pKa = 10.56DD24 pKa = 4.52VFNICLKK31 pKa = 10.88FIMKK35 pKa = 8.68PHH37 pKa = 6.52ILIMYY42 pKa = 8.08ACLLVVVYY50 pKa = 10.65NKK52 pKa = 10.92GEE54 pKa = 4.16AGDD57 pKa = 4.18VIEE60 pKa = 5.21KK61 pKa = 10.4ALKK64 pKa = 9.9AYY66 pKa = 8.41PRR68 pKa = 11.84NPILKK73 pKa = 8.56WAKK76 pKa = 7.64TDD78 pKa = 3.36YY79 pKa = 11.03VRR81 pKa = 11.84FIGILVGLPVIIDD94 pKa = 3.74APRR97 pKa = 11.84SFQLLITGAVFVSTYY112 pKa = 10.31ILPPQTLSTYY122 pKa = 10.92LLGFVSLHH130 pKa = 6.17VYY132 pKa = 10.5SKK134 pKa = 11.12AKK136 pKa = 10.07SVQLRR141 pKa = 11.84IILLASFATYY151 pKa = 10.49LVLSGIVDD159 pKa = 3.71TEE161 pKa = 4.13KK162 pKa = 10.23FTGSGTVLGGAHH174 pKa = 7.3GAPKK178 pKa = 10.21LSEE181 pKa = 4.15VPIPTVLNDD190 pKa = 3.84PGPRR194 pKa = 11.84IRR196 pKa = 11.84SVEE199 pKa = 3.64EE200 pKa = 3.41RR201 pKa = 11.84LPVIKK206 pKa = 10.32RR207 pKa = 11.84GGSGGVV213 pKa = 3.02
Molecular weight: 23.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4327 |
47 |
2645 |
540.9 |
60.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.078 ± 0.571 | 2.843 ± 0.148 |
6.61 ± 0.989 | 4.923 ± 0.47 |
5.778 ± 0.907 | 6.517 ± 0.426 |
2.242 ± 0.47 | 4.761 ± 0.467 |
5.177 ± 0.945 | 9.591 ± 0.955 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.75 ± 0.248 | 3.028 ± 0.302 |
3.975 ± 0.369 | 1.849 ± 0.267 |
6.24 ± 0.592 | 7.788 ± 0.441 |
4.368 ± 0.222 | 10.192 ± 0.447 |
0.878 ± 0.295 | 4.414 ± 0.357 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
