
Glycomyces buryatensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Glycomycetales; Glycomycetaceae; Glycomyces
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
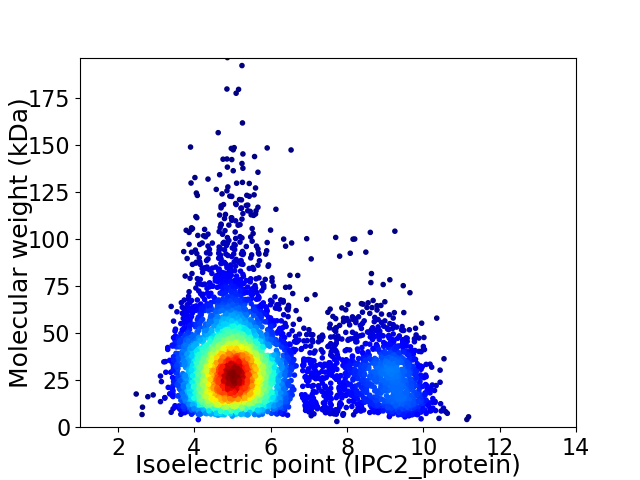
Virtual 2D-PAGE plot for 5187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S8QEG2|A0A4S8QEG2_9ACTN Uncharacterized protein OS=Glycomyces buryatensis OX=2570927 GN=FAB82_03245 PE=4 SV=1
MM1 pKa = 6.89QVHH4 pKa = 6.52RR5 pKa = 11.84SRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.51LIVGVAVASLAASAVTLASANSAFAEE35 pKa = 4.53EE36 pKa = 5.27GCQVDD41 pKa = 4.02YY42 pKa = 11.46NITSSWNGGFQANVVISADD61 pKa = 3.53EE62 pKa = 5.08AINGWDD68 pKa = 3.75VSWDD72 pKa = 3.94FPSGTSVASAWNVDD86 pKa = 3.08WSQSGTTFTGSDD98 pKa = 3.62VGWNASISSGQSRR111 pKa = 11.84EE112 pKa = 3.73VFGFIGSGSSTAPAEE127 pKa = 3.99FTLNGDD133 pKa = 3.61VCDD136 pKa = 4.89GPGTGPTEE144 pKa = 4.49PPTSDD149 pKa = 3.64EE150 pKa = 4.32PTDD153 pKa = 4.04EE154 pKa = 4.49PTDD157 pKa = 4.05PPDD160 pKa = 3.64GSRR163 pKa = 11.84VDD165 pKa = 3.56NPYY168 pKa = 10.68VGAQVYY174 pKa = 8.76VNPTWAANAEE184 pKa = 4.2ASGGEE189 pKa = 4.06AVADD193 pKa = 3.68QPTGVWMDD201 pKa = 4.62RR202 pKa = 11.84ISAIEE207 pKa = 4.44GNDD210 pKa = 3.47SPTTGSMGMRR220 pKa = 11.84DD221 pKa = 4.1HH222 pKa = 7.43LDD224 pKa = 3.28AALTQGADD232 pKa = 4.09LIQFVIYY239 pKa = 10.0NLPGRR244 pKa = 11.84DD245 pKa = 3.5CAALASNGEE254 pKa = 4.21LGAGEE259 pKa = 3.65IDD261 pKa = 3.55RR262 pKa = 11.84YY263 pKa = 9.4KK264 pKa = 11.01NEE266 pKa = 4.36YY267 pKa = 9.03IDD269 pKa = 5.2PIVEE273 pKa = 4.15IQSDD277 pKa = 3.71PAYY280 pKa = 10.81ADD282 pKa = 3.04IKK284 pKa = 9.73IVNVIEE290 pKa = 4.5IDD292 pKa = 3.68SLPNLVTNVSPRR304 pKa = 11.84EE305 pKa = 3.97TATTNCDD312 pKa = 2.82TMLANGNYY320 pKa = 9.54EE321 pKa = 4.46DD322 pKa = 4.12GVSYY326 pKa = 10.74AVSEE330 pKa = 5.36LGDD333 pKa = 3.5QPNSYY338 pKa = 10.55QYY340 pKa = 11.47LDD342 pKa = 4.11AGHH345 pKa = 6.97HH346 pKa = 5.78GWIGWGDD353 pKa = 3.68TNAMYY358 pKa = 11.1DD359 pKa = 3.58NFFASADD366 pKa = 3.78LFGSLIGQNGMTANDD381 pKa = 3.23VSGFITNTANYY392 pKa = 9.28SALEE396 pKa = 3.94EE397 pKa = 4.81PYY399 pKa = 10.61WNVDD403 pKa = 3.66DD404 pKa = 5.05NVGGQAINQNPDD416 pKa = 3.45VKK418 pKa = 10.42WVDD421 pKa = 3.06WNDD424 pKa = 3.45FNGEE428 pKa = 3.98LDD430 pKa = 3.58FAQAFRR436 pKa = 11.84TEE438 pKa = 4.35LVSAGYY444 pKa = 10.63NDD446 pKa = 5.63DD447 pKa = 3.51IGMLIDD453 pKa = 3.64TSRR456 pKa = 11.84NGWGGDD462 pKa = 3.42YY463 pKa = 10.62RR464 pKa = 11.84PTGTSTSTDD473 pKa = 2.82PSTYY477 pKa = 9.87VDD479 pKa = 3.84EE480 pKa = 4.64SRR482 pKa = 11.84IDD484 pKa = 3.27QRR486 pKa = 11.84YY487 pKa = 8.56QKK489 pKa = 11.16GNWCNQAGAGLGEE502 pKa = 4.45RR503 pKa = 11.84PVAAPEE509 pKa = 4.15PGIDD513 pKa = 3.05AYY515 pKa = 11.48VWIKK519 pKa = 10.65PPGEE523 pKa = 3.84SDD525 pKa = 3.57GASEE529 pKa = 6.26LIDD532 pKa = 3.66NDD534 pKa = 3.32EE535 pKa = 4.37GKK537 pKa = 10.97GFDD540 pKa = 3.97EE541 pKa = 4.69MCC543 pKa = 4.8
MM1 pKa = 6.89QVHH4 pKa = 6.52RR5 pKa = 11.84SRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.51LIVGVAVASLAASAVTLASANSAFAEE35 pKa = 4.53EE36 pKa = 5.27GCQVDD41 pKa = 4.02YY42 pKa = 11.46NITSSWNGGFQANVVISADD61 pKa = 3.53EE62 pKa = 5.08AINGWDD68 pKa = 3.75VSWDD72 pKa = 3.94FPSGTSVASAWNVDD86 pKa = 3.08WSQSGTTFTGSDD98 pKa = 3.62VGWNASISSGQSRR111 pKa = 11.84EE112 pKa = 3.73VFGFIGSGSSTAPAEE127 pKa = 3.99FTLNGDD133 pKa = 3.61VCDD136 pKa = 4.89GPGTGPTEE144 pKa = 4.49PPTSDD149 pKa = 3.64EE150 pKa = 4.32PTDD153 pKa = 4.04EE154 pKa = 4.49PTDD157 pKa = 4.05PPDD160 pKa = 3.64GSRR163 pKa = 11.84VDD165 pKa = 3.56NPYY168 pKa = 10.68VGAQVYY174 pKa = 8.76VNPTWAANAEE184 pKa = 4.2ASGGEE189 pKa = 4.06AVADD193 pKa = 3.68QPTGVWMDD201 pKa = 4.62RR202 pKa = 11.84ISAIEE207 pKa = 4.44GNDD210 pKa = 3.47SPTTGSMGMRR220 pKa = 11.84DD221 pKa = 4.1HH222 pKa = 7.43LDD224 pKa = 3.28AALTQGADD232 pKa = 4.09LIQFVIYY239 pKa = 10.0NLPGRR244 pKa = 11.84DD245 pKa = 3.5CAALASNGEE254 pKa = 4.21LGAGEE259 pKa = 3.65IDD261 pKa = 3.55RR262 pKa = 11.84YY263 pKa = 9.4KK264 pKa = 11.01NEE266 pKa = 4.36YY267 pKa = 9.03IDD269 pKa = 5.2PIVEE273 pKa = 4.15IQSDD277 pKa = 3.71PAYY280 pKa = 10.81ADD282 pKa = 3.04IKK284 pKa = 9.73IVNVIEE290 pKa = 4.5IDD292 pKa = 3.68SLPNLVTNVSPRR304 pKa = 11.84EE305 pKa = 3.97TATTNCDD312 pKa = 2.82TMLANGNYY320 pKa = 9.54EE321 pKa = 4.46DD322 pKa = 4.12GVSYY326 pKa = 10.74AVSEE330 pKa = 5.36LGDD333 pKa = 3.5QPNSYY338 pKa = 10.55QYY340 pKa = 11.47LDD342 pKa = 4.11AGHH345 pKa = 6.97HH346 pKa = 5.78GWIGWGDD353 pKa = 3.68TNAMYY358 pKa = 11.1DD359 pKa = 3.58NFFASADD366 pKa = 3.78LFGSLIGQNGMTANDD381 pKa = 3.23VSGFITNTANYY392 pKa = 9.28SALEE396 pKa = 3.94EE397 pKa = 4.81PYY399 pKa = 10.61WNVDD403 pKa = 3.66DD404 pKa = 5.05NVGGQAINQNPDD416 pKa = 3.45VKK418 pKa = 10.42WVDD421 pKa = 3.06WNDD424 pKa = 3.45FNGEE428 pKa = 3.98LDD430 pKa = 3.58FAQAFRR436 pKa = 11.84TEE438 pKa = 4.35LVSAGYY444 pKa = 10.63NDD446 pKa = 5.63DD447 pKa = 3.51IGMLIDD453 pKa = 3.64TSRR456 pKa = 11.84NGWGGDD462 pKa = 3.42YY463 pKa = 10.62RR464 pKa = 11.84PTGTSTSTDD473 pKa = 2.82PSTYY477 pKa = 9.87VDD479 pKa = 3.84EE480 pKa = 4.64SRR482 pKa = 11.84IDD484 pKa = 3.27QRR486 pKa = 11.84YY487 pKa = 8.56QKK489 pKa = 11.16GNWCNQAGAGLGEE502 pKa = 4.45RR503 pKa = 11.84PVAAPEE509 pKa = 4.15PGIDD513 pKa = 3.05AYY515 pKa = 11.48VWIKK519 pKa = 10.65PPGEE523 pKa = 3.84SDD525 pKa = 3.57GASEE529 pKa = 6.26LIDD532 pKa = 3.66NDD534 pKa = 3.32EE535 pKa = 4.37GKK537 pKa = 10.97GFDD540 pKa = 3.97EE541 pKa = 4.69MCC543 pKa = 4.8
Molecular weight: 57.81 kDa
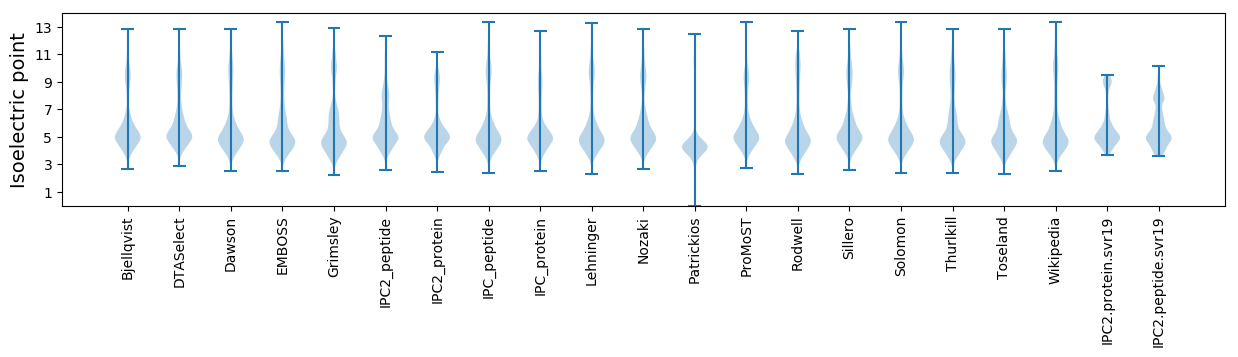
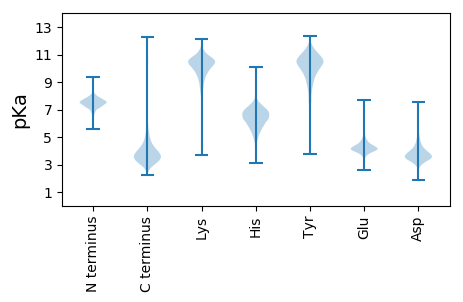
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V4HQV1|A0A4V4HQV1_9ACTN Amidohydrolase OS=Glycomyces buryatensis OX=2570927 GN=FAB82_23520 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1676623 |
29 |
1801 |
323.2 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.87 ± 0.048 | 0.783 ± 0.01 |
6.464 ± 0.032 | 6.546 ± 0.038 |
3.103 ± 0.018 | 8.926 ± 0.032 |
2.111 ± 0.016 | 4.259 ± 0.024 |
2.311 ± 0.023 | 9.757 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.956 ± 0.015 | 2.279 ± 0.021 |
5.461 ± 0.032 | 2.881 ± 0.018 |
7.083 ± 0.041 | 5.508 ± 0.023 |
5.905 ± 0.025 | 7.816 ± 0.029 |
1.707 ± 0.016 | 2.274 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
