
Andreprevotia lacus DSM 23236
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Neisseriales; Chromobacteriaceae; Andreprevotia; Andreprevotia lacus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
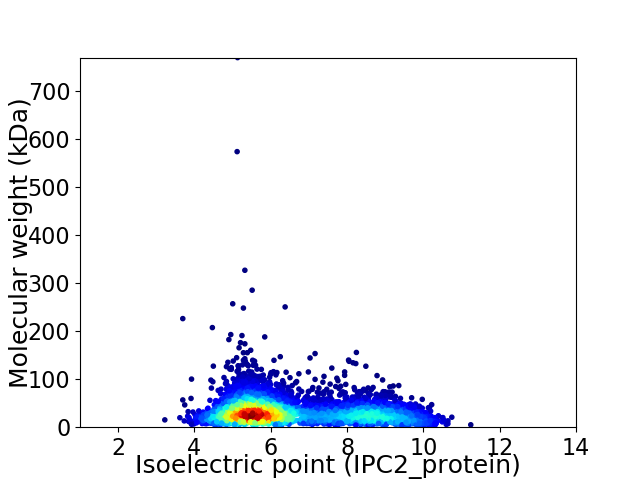
Virtual 2D-PAGE plot for 4303 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W1Y0Q2|A0A1W1Y0Q2_9NEIS Multidrug efflux pump OS=Andreprevotia lacus DSM 23236 OX=1121001 GN=SAMN02745857_04055 PE=4 SV=1
MM1 pKa = 7.47ATPSQQATIVQLNGQAFIRR20 pKa = 11.84DD21 pKa = 3.92ATGKK25 pKa = 7.27MTPLHH30 pKa = 6.46SGEE33 pKa = 4.78TLNQDD38 pKa = 3.07QVLVTSPDD46 pKa = 3.48GEE48 pKa = 4.5VVVLLPNGDD57 pKa = 3.83TQTVGPDD64 pKa = 2.87RR65 pKa = 11.84SVLLDD70 pKa = 3.57AQFMGAAPDD79 pKa = 3.63TTEE82 pKa = 3.98AAVAPSSVQGGTAQIDD98 pKa = 3.57QIVAQGGDD106 pKa = 3.28LSTDD110 pKa = 3.94LEE112 pKa = 4.22ATAAGLTGGGDD123 pKa = 3.61AGNGTHH129 pKa = 5.71TFIRR133 pKa = 11.84VGRR136 pKa = 11.84ISEE139 pKa = 4.37EE140 pKa = 3.94AGSTAFGTPGTPIGGAGAAEE160 pKa = 3.96QNGQNHH166 pKa = 6.39APVAVDD172 pKa = 4.31DD173 pKa = 4.67NSTGAANDD181 pKa = 3.99GLTSPEE187 pKa = 4.24DD188 pKa = 3.78TPLTISPLLLLSNDD202 pKa = 3.8GDD204 pKa = 4.27PDD206 pKa = 4.4GDD208 pKa = 3.97PLTIVSVQDD217 pKa = 3.69PVHH220 pKa = 5.86GTLSIDD226 pKa = 3.34ANNNIVFTPDD236 pKa = 3.08PNYY239 pKa = 10.8NGPASFSYY247 pKa = 10.47TIDD250 pKa = 3.62DD251 pKa = 4.05GRR253 pKa = 11.84GGTATATVNLNITPVNDD270 pKa = 3.72PATVSNDD277 pKa = 2.8AGTVKK282 pKa = 10.47EE283 pKa = 4.3DD284 pKa = 3.61TPTQTTVGGTLTITDD299 pKa = 3.44VDD301 pKa = 3.43TGEE304 pKa = 4.12QHH306 pKa = 6.82VVAQNVSNPYY316 pKa = 8.87GTFTVDD322 pKa = 5.17ADD324 pKa = 4.4GNWSFTIDD332 pKa = 3.46NTSPTVQALAEE343 pKa = 4.38GQTQVLTFDD352 pKa = 3.67VTSVDD357 pKa = 3.41GTGTGVVTITVVGTNDD373 pKa = 3.14NAVITAHH380 pKa = 6.98LPGDD384 pKa = 3.63DD385 pKa = 3.67AGTVKK390 pKa = 10.61EE391 pKa = 4.2DD392 pKa = 3.86TILATGGKK400 pKa = 9.7LDD402 pKa = 4.3VADD405 pKa = 5.15PDD407 pKa = 3.94TGEE410 pKa = 4.26SAVIAQTDD418 pKa = 3.53TTGTYY423 pKa = 10.83GSFSILADD431 pKa = 3.74GTWTYY436 pKa = 9.98TLNNASPVVQALNEE450 pKa = 4.4GEE452 pKa = 4.18SRR454 pKa = 11.84TEE456 pKa = 3.78VFQVQSQDD464 pKa = 2.92GSDD467 pKa = 3.21THH469 pKa = 6.62FVTVTVLGTNDD480 pKa = 3.21NAVITAHH487 pKa = 6.98LPGDD491 pKa = 3.63DD492 pKa = 3.67AGTVKK497 pKa = 10.61EE498 pKa = 4.24DD499 pKa = 4.14TILSTGGKK507 pKa = 9.71LDD509 pKa = 3.81VADD512 pKa = 4.76PDD514 pKa = 3.94AGQSAVVAQSNTTGTYY530 pKa = 10.27GSFSIAADD538 pKa = 3.93GTWTYY543 pKa = 11.53SLNNGSPIVQALNEE557 pKa = 4.42GEE559 pKa = 4.37SRR561 pKa = 11.84TEE563 pKa = 3.84TFQVQSLDD571 pKa = 3.4GTDD574 pKa = 3.14THH576 pKa = 6.4TVTVTILGTNDD587 pKa = 2.93AALIGGNDD595 pKa = 3.42SGTVKK600 pKa = 10.68EE601 pKa = 4.5DD602 pKa = 3.31TTLVTGGILTVSDD615 pKa = 3.57VDD617 pKa = 3.74TGQSSLVAEE626 pKa = 4.72TVTGTYY632 pKa = 8.95GTFSVDD638 pKa = 4.21ANGNWSYY645 pKa = 11.82VLNNSSPIVQALNDD659 pKa = 3.43GDD661 pKa = 4.05TRR663 pKa = 11.84VEE665 pKa = 4.16TFAVQSVDD673 pKa = 3.3GTTHH677 pKa = 5.92TVTVNVLGTNDD688 pKa = 4.21GEE690 pKa = 4.34QLPPPRR696 pKa = 11.84ISVSLDD702 pKa = 3.24SEE704 pKa = 4.36GDD706 pKa = 3.69TVFEE710 pKa = 3.85GNYY713 pKa = 10.63AVFNVHH719 pKa = 6.77LSYY722 pKa = 10.97PSTTDD727 pKa = 3.01VTFTPSQVSGTATVGTDD744 pKa = 3.05TSDD747 pKa = 3.18TLEE750 pKa = 4.13YY751 pKa = 9.98WNGSAWVAVSGGITIPAGQTDD772 pKa = 3.84VQVRR776 pKa = 11.84VATVADD782 pKa = 5.15GIYY785 pKa = 10.2EE786 pKa = 4.17GNEE789 pKa = 3.81NFSLHH794 pKa = 6.36VDD796 pKa = 3.56VTSGSTSNTSADD808 pKa = 3.39ATAVIIDD815 pKa = 3.71GDD817 pKa = 4.0KK818 pKa = 10.62PPEE821 pKa = 3.87VSISVSGEE829 pKa = 3.08GSMVHH834 pKa = 6.17EE835 pKa = 4.48GDD837 pKa = 3.58YY838 pKa = 11.46AVFNVHH844 pKa = 6.73LSGSSSTDD852 pKa = 3.22VVFTPSLVSGTATVGTDD869 pKa = 2.5TGTALEE875 pKa = 4.19YY876 pKa = 11.0SLNGTDD882 pKa = 3.08WFAVGTGVTIPAGQTDD898 pKa = 3.84VQVRR902 pKa = 11.84VATVADD908 pKa = 5.15GIYY911 pKa = 10.2EE912 pKa = 4.17GNEE915 pKa = 3.81NFSLHH920 pKa = 6.47VDD922 pKa = 3.54VTSSNTSNSSADD934 pKa = 3.29ATATIVDD941 pKa = 4.34ADD943 pKa = 4.28SPPQVTVSVSGEE955 pKa = 3.34GSMVHH960 pKa = 6.17EE961 pKa = 4.48GDD963 pKa = 3.58YY964 pKa = 11.46AVFNVHH970 pKa = 6.73LSGSSSTDD978 pKa = 3.22VVFTPSLVSGTATVGTDD995 pKa = 2.5TGTALEE1001 pKa = 4.3YY1002 pKa = 10.23WNGSAWVAISGGVTIPAGQTDD1023 pKa = 3.84VQVRR1027 pKa = 11.84VATVADD1033 pKa = 5.15GIYY1036 pKa = 10.2EE1037 pKa = 4.17GNEE1040 pKa = 3.81NFSLHH1045 pKa = 6.47VDD1047 pKa = 3.54VTSSNTSNTSADD1059 pKa = 3.31ATATIIDD1066 pKa = 4.17ADD1068 pKa = 4.1SPPVVSVSVSSEE1080 pKa = 3.22GSMVPEE1086 pKa = 4.12GAYY1089 pKa = 10.47AVFNVHH1095 pKa = 6.84LSGPSSGAISFTPSLHH1111 pKa = 7.25DD1112 pKa = 3.2GTAIIGTDD1120 pKa = 2.95TSTTLEE1126 pKa = 4.2YY1127 pKa = 10.42WNGSAWTVVSGDD1139 pKa = 2.95ITLAAGQTDD1148 pKa = 3.77VQVRR1152 pKa = 11.84VATIDD1157 pKa = 3.43DD1158 pKa = 4.24SIYY1161 pKa = 10.71EE1162 pKa = 4.16GNEE1165 pKa = 3.65NFSLQVHH1172 pKa = 5.22VTSGSTSNTDD1182 pKa = 3.0ASATAIIVDD1191 pKa = 3.96NDD1193 pKa = 3.72AQPVGHH1199 pKa = 7.59DD1200 pKa = 3.8DD1201 pKa = 4.16GYY1203 pKa = 9.47TVQEE1207 pKa = 4.07ASTNALASVLGNDD1220 pKa = 4.02TPSGLHH1226 pKa = 4.49VAKK1229 pKa = 10.21FATDD1233 pKa = 3.6GAGGGAVSADD1243 pKa = 3.23GSHH1246 pKa = 7.48TITTSLGGTVTMHH1259 pKa = 7.51ADD1261 pKa = 3.22GTFTYY1266 pKa = 8.65TIPDD1270 pKa = 3.54LHH1272 pKa = 6.46HH1273 pKa = 6.98TGTAVDD1279 pKa = 3.52SFYY1282 pKa = 11.59YY1283 pKa = 10.12QATDD1287 pKa = 3.61GNANTAWTKK1296 pKa = 10.47VSVNVTDD1303 pKa = 4.07TGPTAHH1309 pKa = 7.81DD1310 pKa = 3.83DD1311 pKa = 3.98TVSLNTVSTDD1321 pKa = 3.15TVNLVLVVDD1330 pKa = 4.33SSGSIGTTNLNTIKK1344 pKa = 10.63AALSHH1349 pKa = 6.8LMTTYY1354 pKa = 10.95GSALQSVMLVDD1365 pKa = 4.91FDD1367 pKa = 5.09DD1368 pKa = 3.98NARR1371 pKa = 11.84VFHH1374 pKa = 6.53VNTDD1378 pKa = 3.28GTGSVWLTQSQAEE1391 pKa = 4.36SKK1393 pKa = 10.49YY1394 pKa = 11.2SSITSGGNTDD1404 pKa = 3.39YY1405 pKa = 11.71DD1406 pKa = 3.94DD1407 pKa = 5.58AIKK1410 pKa = 10.43AVQDD1414 pKa = 3.58NYY1416 pKa = 9.93GTPPTAAKK1424 pKa = 9.0TYY1426 pKa = 10.52VYY1428 pKa = 9.72FVSDD1432 pKa = 4.2GEE1434 pKa = 4.41PTGSDD1439 pKa = 2.92GTNPNTIEE1447 pKa = 4.0PSEE1450 pKa = 4.02RR1451 pKa = 11.84ADD1453 pKa = 2.88WVNFLTGKK1461 pKa = 9.72VDD1463 pKa = 3.32EE1464 pKa = 5.21VYY1466 pKa = 10.77AIGIGSGVSQTDD1478 pKa = 3.0SDD1480 pKa = 4.51LLDD1483 pKa = 3.87VAWSSSGNDD1492 pKa = 3.17AGNVKK1497 pKa = 10.41LISTADD1503 pKa = 3.88DD1504 pKa = 3.72LSGTLTSIAQSNGNVTSNDD1523 pKa = 3.35DD1524 pKa = 3.63FGLDD1528 pKa = 3.56GAGSPKK1534 pKa = 10.19LVSVSYY1540 pKa = 10.73DD1541 pKa = 3.26ADD1543 pKa = 3.72GSGPGAATVYY1553 pKa = 10.33TFDD1556 pKa = 4.13ATHH1559 pKa = 6.46HH1560 pKa = 6.89AYY1562 pKa = 9.39TINLGAGRR1570 pKa = 11.84GTLVLHH1576 pKa = 7.61DD1577 pKa = 5.29DD1578 pKa = 3.63GSYY1581 pKa = 10.19TYY1583 pKa = 9.75TPASGSSNGAPFTVQYY1599 pKa = 9.46TIQDD1603 pKa = 3.52ADD1605 pKa = 3.96GSQSTASMLFDD1616 pKa = 4.66PNGTPVVGTAPSITVSEE1633 pKa = 4.32EE1634 pKa = 3.85GLPGANPDD1642 pKa = 3.57TTGNTDD1648 pKa = 3.34TTNLALRR1655 pKa = 11.84SGTIAITDD1663 pKa = 3.82PDD1665 pKa = 3.81STDD1668 pKa = 3.38FNLRR1672 pKa = 11.84FAAPTATLTSGGTALSWSGAGTGVLIGMANGVEE1705 pKa = 4.81IIRR1708 pKa = 11.84ISQSDD1713 pKa = 3.41THH1715 pKa = 7.53GGYY1718 pKa = 9.33QVQLSGSLDD1727 pKa = 3.42HH1728 pKa = 6.91ATAGQEE1734 pKa = 4.09DD1735 pKa = 4.61VKK1737 pKa = 11.41SFGVLVTVDD1746 pKa = 4.48DD1747 pKa = 4.5GAAANNTTTTTLTVNVEE1764 pKa = 4.11DD1765 pKa = 5.19DD1766 pKa = 3.92SPINMITNGLAQNTTSTSFEE1786 pKa = 3.92GTLASIGADD1795 pKa = 2.79KK1796 pKa = 10.69TGAHH1800 pKa = 6.15IVLSGSVPSGLTSNGVAITYY1820 pKa = 9.82SLSSDD1825 pKa = 3.26GSTLTAKK1832 pKa = 10.71AGTNTVFTLTGHH1844 pKa = 7.23ADD1846 pKa = 3.02GSYY1849 pKa = 10.65SYY1851 pKa = 11.28DD1852 pKa = 3.25QVRR1855 pKa = 11.84TLDD1858 pKa = 3.76LSVVTSNLQSSVGAGGPQAAYY1879 pKa = 11.2YY1880 pKa = 9.06MYY1882 pKa = 10.08TDD1884 pKa = 4.02GSFGSTEE1891 pKa = 3.95NAKK1894 pKa = 10.44DD1895 pKa = 3.25WAVKK1899 pKa = 8.75ITGSDD1904 pKa = 3.74SINPSTQGMGVSNNLFQTGEE1924 pKa = 3.97TMRR1927 pKa = 11.84FEE1929 pKa = 5.0FDD1931 pKa = 4.14DD1932 pKa = 3.74EE1933 pKa = 4.4HH1934 pKa = 8.05ASTVGGSTPNLAYY1947 pKa = 8.89ATKK1950 pKa = 10.32IGVTDD1955 pKa = 3.75LTGSEE1960 pKa = 4.14SLAWTAYY1967 pKa = 8.54FTDD1970 pKa = 4.8GSHH1973 pKa = 7.14ASGVVTAGSLVSGAFTITSPNGLYY1997 pKa = 10.38LDD1999 pKa = 4.36YY2000 pKa = 11.51VDD2002 pKa = 5.43LAPGSSTSIRR2012 pKa = 11.84LTSVTNYY2019 pKa = 10.35VLDD2022 pKa = 4.1DD2023 pKa = 3.68THH2025 pKa = 7.11TKK2027 pKa = 10.56DD2028 pKa = 4.23LNFGFTAYY2036 pKa = 10.43DD2037 pKa = 3.65GDD2039 pKa = 4.28GDD2041 pKa = 4.08TSNGSLTITSQNSATLTGDD2060 pKa = 3.32ATHH2063 pKa = 6.85TALGGGNANNTLLGTAGNDD2082 pKa = 2.95ILTGGAGNDD2091 pKa = 3.65TLTGGLGADD2100 pKa = 3.53VFRR2103 pKa = 11.84WGVNDD2108 pKa = 4.11HH2109 pKa = 5.97GTAATPSNDD2118 pKa = 2.9VVTDD2122 pKa = 4.05FATTGASHH2130 pKa = 7.28DD2131 pKa = 3.9QLDD2134 pKa = 4.28LRR2136 pKa = 11.84DD2137 pKa = 4.51LLTSVNWTVNAGHH2150 pKa = 6.22TTAQLATEE2158 pKa = 4.33LDD2160 pKa = 4.25GYY2162 pKa = 11.21LSVSEE2167 pKa = 4.72SGTTTTLSVKK2177 pKa = 10.28AGSSSVTEE2185 pKa = 4.6TIQLQNVTDD2194 pKa = 4.33LVDD2197 pKa = 3.72GTHH2200 pKa = 5.8TVSHH2204 pKa = 6.51DD2205 pKa = 3.74VIVYY2209 pKa = 9.14MLDD2212 pKa = 3.2HH2213 pKa = 7.4DD2214 pKa = 5.68LIKK2217 pKa = 10.61HH2218 pKa = 5.95DD2219 pKa = 3.93
MM1 pKa = 7.47ATPSQQATIVQLNGQAFIRR20 pKa = 11.84DD21 pKa = 3.92ATGKK25 pKa = 7.27MTPLHH30 pKa = 6.46SGEE33 pKa = 4.78TLNQDD38 pKa = 3.07QVLVTSPDD46 pKa = 3.48GEE48 pKa = 4.5VVVLLPNGDD57 pKa = 3.83TQTVGPDD64 pKa = 2.87RR65 pKa = 11.84SVLLDD70 pKa = 3.57AQFMGAAPDD79 pKa = 3.63TTEE82 pKa = 3.98AAVAPSSVQGGTAQIDD98 pKa = 3.57QIVAQGGDD106 pKa = 3.28LSTDD110 pKa = 3.94LEE112 pKa = 4.22ATAAGLTGGGDD123 pKa = 3.61AGNGTHH129 pKa = 5.71TFIRR133 pKa = 11.84VGRR136 pKa = 11.84ISEE139 pKa = 4.37EE140 pKa = 3.94AGSTAFGTPGTPIGGAGAAEE160 pKa = 3.96QNGQNHH166 pKa = 6.39APVAVDD172 pKa = 4.31DD173 pKa = 4.67NSTGAANDD181 pKa = 3.99GLTSPEE187 pKa = 4.24DD188 pKa = 3.78TPLTISPLLLLSNDD202 pKa = 3.8GDD204 pKa = 4.27PDD206 pKa = 4.4GDD208 pKa = 3.97PLTIVSVQDD217 pKa = 3.69PVHH220 pKa = 5.86GTLSIDD226 pKa = 3.34ANNNIVFTPDD236 pKa = 3.08PNYY239 pKa = 10.8NGPASFSYY247 pKa = 10.47TIDD250 pKa = 3.62DD251 pKa = 4.05GRR253 pKa = 11.84GGTATATVNLNITPVNDD270 pKa = 3.72PATVSNDD277 pKa = 2.8AGTVKK282 pKa = 10.47EE283 pKa = 4.3DD284 pKa = 3.61TPTQTTVGGTLTITDD299 pKa = 3.44VDD301 pKa = 3.43TGEE304 pKa = 4.12QHH306 pKa = 6.82VVAQNVSNPYY316 pKa = 8.87GTFTVDD322 pKa = 5.17ADD324 pKa = 4.4GNWSFTIDD332 pKa = 3.46NTSPTVQALAEE343 pKa = 4.38GQTQVLTFDD352 pKa = 3.67VTSVDD357 pKa = 3.41GTGTGVVTITVVGTNDD373 pKa = 3.14NAVITAHH380 pKa = 6.98LPGDD384 pKa = 3.63DD385 pKa = 3.67AGTVKK390 pKa = 10.61EE391 pKa = 4.2DD392 pKa = 3.86TILATGGKK400 pKa = 9.7LDD402 pKa = 4.3VADD405 pKa = 5.15PDD407 pKa = 3.94TGEE410 pKa = 4.26SAVIAQTDD418 pKa = 3.53TTGTYY423 pKa = 10.83GSFSILADD431 pKa = 3.74GTWTYY436 pKa = 9.98TLNNASPVVQALNEE450 pKa = 4.4GEE452 pKa = 4.18SRR454 pKa = 11.84TEE456 pKa = 3.78VFQVQSQDD464 pKa = 2.92GSDD467 pKa = 3.21THH469 pKa = 6.62FVTVTVLGTNDD480 pKa = 3.21NAVITAHH487 pKa = 6.98LPGDD491 pKa = 3.63DD492 pKa = 3.67AGTVKK497 pKa = 10.61EE498 pKa = 4.24DD499 pKa = 4.14TILSTGGKK507 pKa = 9.71LDD509 pKa = 3.81VADD512 pKa = 4.76PDD514 pKa = 3.94AGQSAVVAQSNTTGTYY530 pKa = 10.27GSFSIAADD538 pKa = 3.93GTWTYY543 pKa = 11.53SLNNGSPIVQALNEE557 pKa = 4.42GEE559 pKa = 4.37SRR561 pKa = 11.84TEE563 pKa = 3.84TFQVQSLDD571 pKa = 3.4GTDD574 pKa = 3.14THH576 pKa = 6.4TVTVTILGTNDD587 pKa = 2.93AALIGGNDD595 pKa = 3.42SGTVKK600 pKa = 10.68EE601 pKa = 4.5DD602 pKa = 3.31TTLVTGGILTVSDD615 pKa = 3.57VDD617 pKa = 3.74TGQSSLVAEE626 pKa = 4.72TVTGTYY632 pKa = 8.95GTFSVDD638 pKa = 4.21ANGNWSYY645 pKa = 11.82VLNNSSPIVQALNDD659 pKa = 3.43GDD661 pKa = 4.05TRR663 pKa = 11.84VEE665 pKa = 4.16TFAVQSVDD673 pKa = 3.3GTTHH677 pKa = 5.92TVTVNVLGTNDD688 pKa = 4.21GEE690 pKa = 4.34QLPPPRR696 pKa = 11.84ISVSLDD702 pKa = 3.24SEE704 pKa = 4.36GDD706 pKa = 3.69TVFEE710 pKa = 3.85GNYY713 pKa = 10.63AVFNVHH719 pKa = 6.77LSYY722 pKa = 10.97PSTTDD727 pKa = 3.01VTFTPSQVSGTATVGTDD744 pKa = 3.05TSDD747 pKa = 3.18TLEE750 pKa = 4.13YY751 pKa = 9.98WNGSAWVAVSGGITIPAGQTDD772 pKa = 3.84VQVRR776 pKa = 11.84VATVADD782 pKa = 5.15GIYY785 pKa = 10.2EE786 pKa = 4.17GNEE789 pKa = 3.81NFSLHH794 pKa = 6.36VDD796 pKa = 3.56VTSGSTSNTSADD808 pKa = 3.39ATAVIIDD815 pKa = 3.71GDD817 pKa = 4.0KK818 pKa = 10.62PPEE821 pKa = 3.87VSISVSGEE829 pKa = 3.08GSMVHH834 pKa = 6.17EE835 pKa = 4.48GDD837 pKa = 3.58YY838 pKa = 11.46AVFNVHH844 pKa = 6.73LSGSSSTDD852 pKa = 3.22VVFTPSLVSGTATVGTDD869 pKa = 2.5TGTALEE875 pKa = 4.19YY876 pKa = 11.0SLNGTDD882 pKa = 3.08WFAVGTGVTIPAGQTDD898 pKa = 3.84VQVRR902 pKa = 11.84VATVADD908 pKa = 5.15GIYY911 pKa = 10.2EE912 pKa = 4.17GNEE915 pKa = 3.81NFSLHH920 pKa = 6.47VDD922 pKa = 3.54VTSSNTSNSSADD934 pKa = 3.29ATATIVDD941 pKa = 4.34ADD943 pKa = 4.28SPPQVTVSVSGEE955 pKa = 3.34GSMVHH960 pKa = 6.17EE961 pKa = 4.48GDD963 pKa = 3.58YY964 pKa = 11.46AVFNVHH970 pKa = 6.73LSGSSSTDD978 pKa = 3.22VVFTPSLVSGTATVGTDD995 pKa = 2.5TGTALEE1001 pKa = 4.3YY1002 pKa = 10.23WNGSAWVAISGGVTIPAGQTDD1023 pKa = 3.84VQVRR1027 pKa = 11.84VATVADD1033 pKa = 5.15GIYY1036 pKa = 10.2EE1037 pKa = 4.17GNEE1040 pKa = 3.81NFSLHH1045 pKa = 6.47VDD1047 pKa = 3.54VTSSNTSNTSADD1059 pKa = 3.31ATATIIDD1066 pKa = 4.17ADD1068 pKa = 4.1SPPVVSVSVSSEE1080 pKa = 3.22GSMVPEE1086 pKa = 4.12GAYY1089 pKa = 10.47AVFNVHH1095 pKa = 6.84LSGPSSGAISFTPSLHH1111 pKa = 7.25DD1112 pKa = 3.2GTAIIGTDD1120 pKa = 2.95TSTTLEE1126 pKa = 4.2YY1127 pKa = 10.42WNGSAWTVVSGDD1139 pKa = 2.95ITLAAGQTDD1148 pKa = 3.77VQVRR1152 pKa = 11.84VATIDD1157 pKa = 3.43DD1158 pKa = 4.24SIYY1161 pKa = 10.71EE1162 pKa = 4.16GNEE1165 pKa = 3.65NFSLQVHH1172 pKa = 5.22VTSGSTSNTDD1182 pKa = 3.0ASATAIIVDD1191 pKa = 3.96NDD1193 pKa = 3.72AQPVGHH1199 pKa = 7.59DD1200 pKa = 3.8DD1201 pKa = 4.16GYY1203 pKa = 9.47TVQEE1207 pKa = 4.07ASTNALASVLGNDD1220 pKa = 4.02TPSGLHH1226 pKa = 4.49VAKK1229 pKa = 10.21FATDD1233 pKa = 3.6GAGGGAVSADD1243 pKa = 3.23GSHH1246 pKa = 7.48TITTSLGGTVTMHH1259 pKa = 7.51ADD1261 pKa = 3.22GTFTYY1266 pKa = 8.65TIPDD1270 pKa = 3.54LHH1272 pKa = 6.46HH1273 pKa = 6.98TGTAVDD1279 pKa = 3.52SFYY1282 pKa = 11.59YY1283 pKa = 10.12QATDD1287 pKa = 3.61GNANTAWTKK1296 pKa = 10.47VSVNVTDD1303 pKa = 4.07TGPTAHH1309 pKa = 7.81DD1310 pKa = 3.83DD1311 pKa = 3.98TVSLNTVSTDD1321 pKa = 3.15TVNLVLVVDD1330 pKa = 4.33SSGSIGTTNLNTIKK1344 pKa = 10.63AALSHH1349 pKa = 6.8LMTTYY1354 pKa = 10.95GSALQSVMLVDD1365 pKa = 4.91FDD1367 pKa = 5.09DD1368 pKa = 3.98NARR1371 pKa = 11.84VFHH1374 pKa = 6.53VNTDD1378 pKa = 3.28GTGSVWLTQSQAEE1391 pKa = 4.36SKK1393 pKa = 10.49YY1394 pKa = 11.2SSITSGGNTDD1404 pKa = 3.39YY1405 pKa = 11.71DD1406 pKa = 3.94DD1407 pKa = 5.58AIKK1410 pKa = 10.43AVQDD1414 pKa = 3.58NYY1416 pKa = 9.93GTPPTAAKK1424 pKa = 9.0TYY1426 pKa = 10.52VYY1428 pKa = 9.72FVSDD1432 pKa = 4.2GEE1434 pKa = 4.41PTGSDD1439 pKa = 2.92GTNPNTIEE1447 pKa = 4.0PSEE1450 pKa = 4.02RR1451 pKa = 11.84ADD1453 pKa = 2.88WVNFLTGKK1461 pKa = 9.72VDD1463 pKa = 3.32EE1464 pKa = 5.21VYY1466 pKa = 10.77AIGIGSGVSQTDD1478 pKa = 3.0SDD1480 pKa = 4.51LLDD1483 pKa = 3.87VAWSSSGNDD1492 pKa = 3.17AGNVKK1497 pKa = 10.41LISTADD1503 pKa = 3.88DD1504 pKa = 3.72LSGTLTSIAQSNGNVTSNDD1523 pKa = 3.35DD1524 pKa = 3.63FGLDD1528 pKa = 3.56GAGSPKK1534 pKa = 10.19LVSVSYY1540 pKa = 10.73DD1541 pKa = 3.26ADD1543 pKa = 3.72GSGPGAATVYY1553 pKa = 10.33TFDD1556 pKa = 4.13ATHH1559 pKa = 6.46HH1560 pKa = 6.89AYY1562 pKa = 9.39TINLGAGRR1570 pKa = 11.84GTLVLHH1576 pKa = 7.61DD1577 pKa = 5.29DD1578 pKa = 3.63GSYY1581 pKa = 10.19TYY1583 pKa = 9.75TPASGSSNGAPFTVQYY1599 pKa = 9.46TIQDD1603 pKa = 3.52ADD1605 pKa = 3.96GSQSTASMLFDD1616 pKa = 4.66PNGTPVVGTAPSITVSEE1633 pKa = 4.32EE1634 pKa = 3.85GLPGANPDD1642 pKa = 3.57TTGNTDD1648 pKa = 3.34TTNLALRR1655 pKa = 11.84SGTIAITDD1663 pKa = 3.82PDD1665 pKa = 3.81STDD1668 pKa = 3.38FNLRR1672 pKa = 11.84FAAPTATLTSGGTALSWSGAGTGVLIGMANGVEE1705 pKa = 4.81IIRR1708 pKa = 11.84ISQSDD1713 pKa = 3.41THH1715 pKa = 7.53GGYY1718 pKa = 9.33QVQLSGSLDD1727 pKa = 3.42HH1728 pKa = 6.91ATAGQEE1734 pKa = 4.09DD1735 pKa = 4.61VKK1737 pKa = 11.41SFGVLVTVDD1746 pKa = 4.48DD1747 pKa = 4.5GAAANNTTTTTLTVNVEE1764 pKa = 4.11DD1765 pKa = 5.19DD1766 pKa = 3.92SPINMITNGLAQNTTSTSFEE1786 pKa = 3.92GTLASIGADD1795 pKa = 2.79KK1796 pKa = 10.69TGAHH1800 pKa = 6.15IVLSGSVPSGLTSNGVAITYY1820 pKa = 9.82SLSSDD1825 pKa = 3.26GSTLTAKK1832 pKa = 10.71AGTNTVFTLTGHH1844 pKa = 7.23ADD1846 pKa = 3.02GSYY1849 pKa = 10.65SYY1851 pKa = 11.28DD1852 pKa = 3.25QVRR1855 pKa = 11.84TLDD1858 pKa = 3.76LSVVTSNLQSSVGAGGPQAAYY1879 pKa = 11.2YY1880 pKa = 9.06MYY1882 pKa = 10.08TDD1884 pKa = 4.02GSFGSTEE1891 pKa = 3.95NAKK1894 pKa = 10.44DD1895 pKa = 3.25WAVKK1899 pKa = 8.75ITGSDD1904 pKa = 3.74SINPSTQGMGVSNNLFQTGEE1924 pKa = 3.97TMRR1927 pKa = 11.84FEE1929 pKa = 5.0FDD1931 pKa = 4.14DD1932 pKa = 3.74EE1933 pKa = 4.4HH1934 pKa = 8.05ASTVGGSTPNLAYY1947 pKa = 8.89ATKK1950 pKa = 10.32IGVTDD1955 pKa = 3.75LTGSEE1960 pKa = 4.14SLAWTAYY1967 pKa = 8.54FTDD1970 pKa = 4.8GSHH1973 pKa = 7.14ASGVVTAGSLVSGAFTITSPNGLYY1997 pKa = 10.38LDD1999 pKa = 4.36YY2000 pKa = 11.51VDD2002 pKa = 5.43LAPGSSTSIRR2012 pKa = 11.84LTSVTNYY2019 pKa = 10.35VLDD2022 pKa = 4.1DD2023 pKa = 3.68THH2025 pKa = 7.11TKK2027 pKa = 10.56DD2028 pKa = 4.23LNFGFTAYY2036 pKa = 10.43DD2037 pKa = 3.65GDD2039 pKa = 4.28GDD2041 pKa = 4.08TSNGSLTITSQNSATLTGDD2060 pKa = 3.32ATHH2063 pKa = 6.85TALGGGNANNTLLGTAGNDD2082 pKa = 2.95ILTGGAGNDD2091 pKa = 3.65TLTGGLGADD2100 pKa = 3.53VFRR2103 pKa = 11.84WGVNDD2108 pKa = 4.11HH2109 pKa = 5.97GTAATPSNDD2118 pKa = 2.9VVTDD2122 pKa = 4.05FATTGASHH2130 pKa = 7.28DD2131 pKa = 3.9QLDD2134 pKa = 4.28LRR2136 pKa = 11.84DD2137 pKa = 4.51LLTSVNWTVNAGHH2150 pKa = 6.22TTAQLATEE2158 pKa = 4.33LDD2160 pKa = 4.25GYY2162 pKa = 11.21LSVSEE2167 pKa = 4.72SGTTTTLSVKK2177 pKa = 10.28AGSSSVTEE2185 pKa = 4.6TIQLQNVTDD2194 pKa = 4.33LVDD2197 pKa = 3.72GTHH2200 pKa = 5.8TVSHH2204 pKa = 6.51DD2205 pKa = 3.74VIVYY2209 pKa = 9.14MLDD2212 pKa = 3.2HH2213 pKa = 7.4DD2214 pKa = 5.68LIKK2217 pKa = 10.61HH2218 pKa = 5.95DD2219 pKa = 3.93
Molecular weight: 226.21 kDa
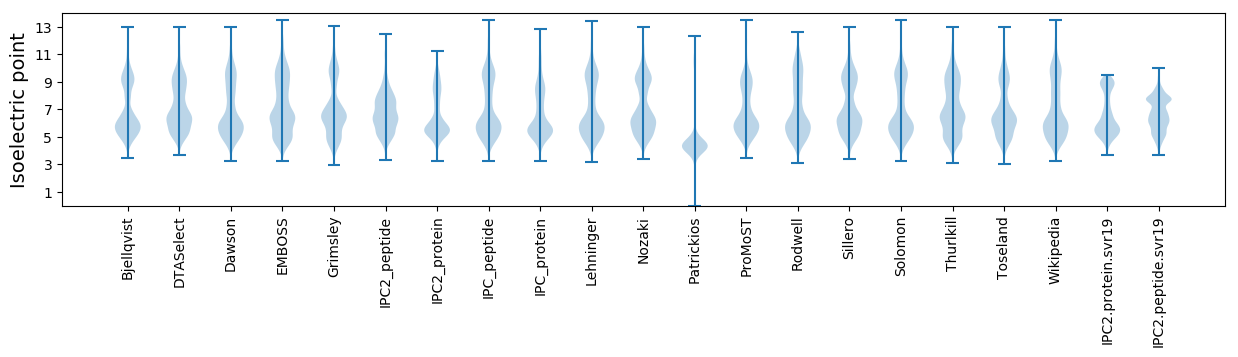
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W1XU88|A0A1W1XU88_9NEIS Membrane fusion protein multidrug efflux system OS=Andreprevotia lacus DSM 23236 OX=1121001 GN=SAMN02745857_02897 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.58RR14 pKa = 11.84THH16 pKa = 6.01GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.58RR14 pKa = 11.84THH16 pKa = 6.01GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12GGRR28 pKa = 11.84RR29 pKa = 11.84VLNARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369701 |
39 |
7105 |
318.3 |
34.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.499 ± 0.053 | 0.933 ± 0.012 |
5.516 ± 0.03 | 4.964 ± 0.04 |
3.454 ± 0.024 | 7.954 ± 0.036 |
2.335 ± 0.021 | 4.531 ± 0.03 |
3.387 ± 0.034 | 11.323 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.162 ± 0.02 | 3.04 ± 0.028 |
5.117 ± 0.034 | 4.651 ± 0.028 |
6.201 ± 0.042 | 5.345 ± 0.035 |
5.289 ± 0.052 | 7.053 ± 0.033 |
1.571 ± 0.02 | 2.675 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
