
Gemmobacter aquatilis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Gemmobacter
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
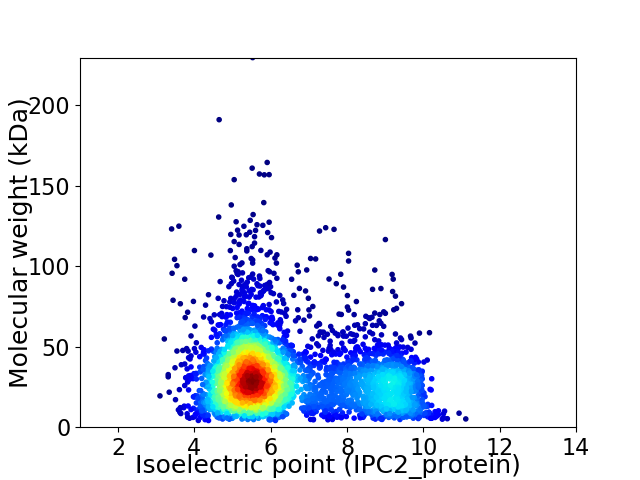
Virtual 2D-PAGE plot for 3845 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8DZF7|A0A1H8DZF7_9RHOB Peptide methionine sulfoxide reductase MsrA OS=Gemmobacter aquatilis OX=933059 GN=msrA PE=3 SV=1
MM1 pKa = 7.4CTLCSALRR9 pKa = 11.84PDD11 pKa = 4.66DD12 pKa = 4.21LAAASDD18 pKa = 3.82LHH20 pKa = 6.32TGAGTGAALPVYY32 pKa = 10.4SLGQIARR39 pKa = 11.84QLTEE43 pKa = 4.97GYY45 pKa = 8.25WAYY48 pKa = 9.87TGRR51 pKa = 11.84DD52 pKa = 3.01MRR54 pKa = 11.84AFAVEE59 pKa = 4.08NGDD62 pKa = 3.73TLTVDD67 pKa = 5.01FGPLSQAEE75 pKa = 4.17RR76 pKa = 11.84AIATAALQAWTDD88 pKa = 3.33VTGITFTAYY97 pKa = 9.56TGDD100 pKa = 4.14DD101 pKa = 3.51PTVIAEE107 pKa = 4.36TTDD110 pKa = 3.3AATGTATTATLPVNGVFSGEE130 pKa = 3.77IDD132 pKa = 3.59STGDD136 pKa = 3.38ADD138 pKa = 4.22WIKK141 pKa = 9.95VTLQKK146 pKa = 10.89GQTYY150 pKa = 10.5RR151 pKa = 11.84ILLEE155 pKa = 4.14SAEE158 pKa = 4.53DD159 pKa = 3.8ADD161 pKa = 5.15ALADD165 pKa = 4.03PLLALHH171 pKa = 7.19DD172 pKa = 3.74ATGAVIATSDD182 pKa = 3.39DD183 pKa = 3.58RR184 pKa = 11.84SRR186 pKa = 11.84GDD188 pKa = 3.52LNSFLSFTAAEE199 pKa = 4.05TGTYY203 pKa = 9.83YY204 pKa = 11.13LSAEE208 pKa = 4.36SATSGSGTYY217 pKa = 11.03YY218 pKa = 9.9MGLVTEE224 pKa = 4.95AAHH227 pKa = 6.41GADD230 pKa = 2.66ISFDD234 pKa = 3.71NRR236 pKa = 11.84DD237 pKa = 3.47SGAYY241 pKa = 9.78AYY243 pKa = 11.01SDD245 pKa = 3.55VAGTRR250 pKa = 11.84ILYY253 pKa = 10.25SYY255 pKa = 11.39VNIARR260 pKa = 11.84NWDD263 pKa = 3.58ASDD266 pKa = 4.75LSLNSYY272 pKa = 8.21WLQTYY277 pKa = 8.66IHH279 pKa = 7.25EE280 pKa = 4.73IGHH283 pKa = 6.42ALGLGHH289 pKa = 7.61AGNYY293 pKa = 6.88NTTASFPTDD302 pKa = 2.54AHH304 pKa = 6.01YY305 pKa = 11.59ANDD308 pKa = 3.71SVTNSIMSYY317 pKa = 9.84FFQGEE322 pKa = 4.25NPNIEE327 pKa = 4.1ADD329 pKa = 3.86FGFLATVMPADD340 pKa = 3.4ILAIQDD346 pKa = 4.08LYY348 pKa = 11.57GSDD351 pKa = 3.65VTTHH355 pKa = 6.96AGDD358 pKa = 3.47TTYY361 pKa = 11.3GANSNVDD368 pKa = 3.47GYY370 pKa = 10.66LGAMFAQIFAEE381 pKa = 4.65APADD385 pKa = 3.42PAVYY389 pKa = 9.91QGEE392 pKa = 4.37NMVLTLYY399 pKa = 9.19DD400 pKa = 3.53TGGHH404 pKa = 6.88DD405 pKa = 6.07LLDD408 pKa = 4.44LSPSRR413 pKa = 11.84GPQVISLLAEE423 pKa = 4.16TFSSIGGYY431 pKa = 10.18LSNLTIARR439 pKa = 11.84GTVIEE444 pKa = 4.68DD445 pKa = 3.66VIAGAGADD453 pKa = 3.59TVLGNAAANMLDD465 pKa = 3.62GAAGSDD471 pKa = 3.49WLIGGAGADD480 pKa = 3.75TLAGGLGNDD489 pKa = 4.25LLKK492 pKa = 11.18GGAGRR497 pKa = 11.84DD498 pKa = 3.43TASFATATRR507 pKa = 11.84GVIVDD512 pKa = 4.63LALTTAQSTGAGQDD526 pKa = 2.85RR527 pKa = 11.84LTGVEE532 pKa = 4.36NLTGSAFNDD541 pKa = 3.6GLGGNGAANVLDD553 pKa = 4.21GVAGRR558 pKa = 11.84DD559 pKa = 3.54LLVGRR564 pKa = 11.84GGNDD568 pKa = 2.88WLLGGAGADD577 pKa = 3.7TLSGGLGRR585 pKa = 11.84DD586 pKa = 3.05AFVFAAGADD595 pKa = 3.66VVLDD599 pKa = 3.82FTDD602 pKa = 5.56DD603 pKa = 3.51EE604 pKa = 5.03DD605 pKa = 4.68SLVFSTALFPGGTTTAEE622 pKa = 3.86EE623 pKa = 4.25LLARR627 pKa = 11.84AAVVSGDD634 pKa = 3.75TVFTFAPGIRR644 pKa = 11.84LILSGFTDD652 pKa = 3.76IAALADD658 pKa = 4.15DD659 pKa = 4.99LLLII663 pKa = 4.93
MM1 pKa = 7.4CTLCSALRR9 pKa = 11.84PDD11 pKa = 4.66DD12 pKa = 4.21LAAASDD18 pKa = 3.82LHH20 pKa = 6.32TGAGTGAALPVYY32 pKa = 10.4SLGQIARR39 pKa = 11.84QLTEE43 pKa = 4.97GYY45 pKa = 8.25WAYY48 pKa = 9.87TGRR51 pKa = 11.84DD52 pKa = 3.01MRR54 pKa = 11.84AFAVEE59 pKa = 4.08NGDD62 pKa = 3.73TLTVDD67 pKa = 5.01FGPLSQAEE75 pKa = 4.17RR76 pKa = 11.84AIATAALQAWTDD88 pKa = 3.33VTGITFTAYY97 pKa = 9.56TGDD100 pKa = 4.14DD101 pKa = 3.51PTVIAEE107 pKa = 4.36TTDD110 pKa = 3.3AATGTATTATLPVNGVFSGEE130 pKa = 3.77IDD132 pKa = 3.59STGDD136 pKa = 3.38ADD138 pKa = 4.22WIKK141 pKa = 9.95VTLQKK146 pKa = 10.89GQTYY150 pKa = 10.5RR151 pKa = 11.84ILLEE155 pKa = 4.14SAEE158 pKa = 4.53DD159 pKa = 3.8ADD161 pKa = 5.15ALADD165 pKa = 4.03PLLALHH171 pKa = 7.19DD172 pKa = 3.74ATGAVIATSDD182 pKa = 3.39DD183 pKa = 3.58RR184 pKa = 11.84SRR186 pKa = 11.84GDD188 pKa = 3.52LNSFLSFTAAEE199 pKa = 4.05TGTYY203 pKa = 9.83YY204 pKa = 11.13LSAEE208 pKa = 4.36SATSGSGTYY217 pKa = 11.03YY218 pKa = 9.9MGLVTEE224 pKa = 4.95AAHH227 pKa = 6.41GADD230 pKa = 2.66ISFDD234 pKa = 3.71NRR236 pKa = 11.84DD237 pKa = 3.47SGAYY241 pKa = 9.78AYY243 pKa = 11.01SDD245 pKa = 3.55VAGTRR250 pKa = 11.84ILYY253 pKa = 10.25SYY255 pKa = 11.39VNIARR260 pKa = 11.84NWDD263 pKa = 3.58ASDD266 pKa = 4.75LSLNSYY272 pKa = 8.21WLQTYY277 pKa = 8.66IHH279 pKa = 7.25EE280 pKa = 4.73IGHH283 pKa = 6.42ALGLGHH289 pKa = 7.61AGNYY293 pKa = 6.88NTTASFPTDD302 pKa = 2.54AHH304 pKa = 6.01YY305 pKa = 11.59ANDD308 pKa = 3.71SVTNSIMSYY317 pKa = 9.84FFQGEE322 pKa = 4.25NPNIEE327 pKa = 4.1ADD329 pKa = 3.86FGFLATVMPADD340 pKa = 3.4ILAIQDD346 pKa = 4.08LYY348 pKa = 11.57GSDD351 pKa = 3.65VTTHH355 pKa = 6.96AGDD358 pKa = 3.47TTYY361 pKa = 11.3GANSNVDD368 pKa = 3.47GYY370 pKa = 10.66LGAMFAQIFAEE381 pKa = 4.65APADD385 pKa = 3.42PAVYY389 pKa = 9.91QGEE392 pKa = 4.37NMVLTLYY399 pKa = 9.19DD400 pKa = 3.53TGGHH404 pKa = 6.88DD405 pKa = 6.07LLDD408 pKa = 4.44LSPSRR413 pKa = 11.84GPQVISLLAEE423 pKa = 4.16TFSSIGGYY431 pKa = 10.18LSNLTIARR439 pKa = 11.84GTVIEE444 pKa = 4.68DD445 pKa = 3.66VIAGAGADD453 pKa = 3.59TVLGNAAANMLDD465 pKa = 3.62GAAGSDD471 pKa = 3.49WLIGGAGADD480 pKa = 3.75TLAGGLGNDD489 pKa = 4.25LLKK492 pKa = 11.18GGAGRR497 pKa = 11.84DD498 pKa = 3.43TASFATATRR507 pKa = 11.84GVIVDD512 pKa = 4.63LALTTAQSTGAGQDD526 pKa = 2.85RR527 pKa = 11.84LTGVEE532 pKa = 4.36NLTGSAFNDD541 pKa = 3.6GLGGNGAANVLDD553 pKa = 4.21GVAGRR558 pKa = 11.84DD559 pKa = 3.54LLVGRR564 pKa = 11.84GGNDD568 pKa = 2.88WLLGGAGADD577 pKa = 3.7TLSGGLGRR585 pKa = 11.84DD586 pKa = 3.05AFVFAAGADD595 pKa = 3.66VVLDD599 pKa = 3.82FTDD602 pKa = 5.56DD603 pKa = 3.51EE604 pKa = 5.03DD605 pKa = 4.68SLVFSTALFPGGTTTAEE622 pKa = 3.86EE623 pKa = 4.25LLARR627 pKa = 11.84AAVVSGDD634 pKa = 3.75TVFTFAPGIRR644 pKa = 11.84LILSGFTDD652 pKa = 3.76IAALADD658 pKa = 4.15DD659 pKa = 4.99LLLII663 pKa = 4.93
Molecular weight: 68.11 kDa
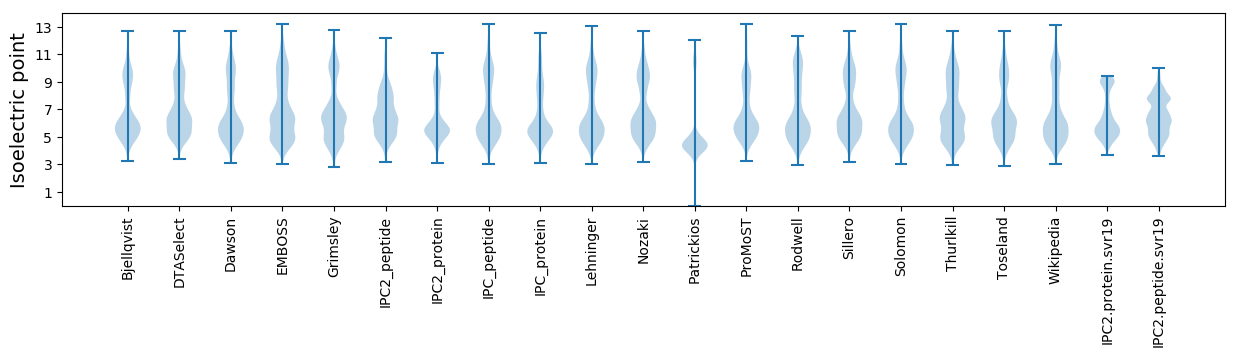
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7ZMX2|A0A1H7ZMX2_9RHOB Thiamine-binding periplasmic protein OS=Gemmobacter aquatilis OX=933059 GN=SAMN04488103_101572 PE=3 SV=1
MM1 pKa = 7.32TRR3 pKa = 11.84FLLILIIGAIAGFLATRR20 pKa = 11.84FMRR23 pKa = 11.84VQTSVGVTVLIGMAGASIAWFVLRR47 pKa = 11.84FVMLLTRR54 pKa = 11.84SGFLLVGAVLGAMALIWLWKK74 pKa = 9.14TFLNRR79 pKa = 3.52
MM1 pKa = 7.32TRR3 pKa = 11.84FLLILIIGAIAGFLATRR20 pKa = 11.84FMRR23 pKa = 11.84VQTSVGVTVLIGMAGASIAWFVLRR47 pKa = 11.84FVMLLTRR54 pKa = 11.84SGFLLVGAVLGAMALIWLWKK74 pKa = 9.14TFLNRR79 pKa = 3.52
Molecular weight: 8.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1190717 |
39 |
2131 |
309.7 |
33.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.69 ± 0.063 | 0.901 ± 0.013 |
5.634 ± 0.034 | 5.485 ± 0.03 |
3.601 ± 0.023 | 9.049 ± 0.042 |
2.056 ± 0.02 | 4.805 ± 0.029 |
2.96 ± 0.033 | 10.632 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.018 | 2.306 ± 0.026 |
5.398 ± 0.04 | 3.187 ± 0.02 |
6.942 ± 0.043 | 4.616 ± 0.025 |
5.322 ± 0.024 | 7.26 ± 0.033 |
1.405 ± 0.016 | 2.083 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
