
Celeribacter baekdonensis B30
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Celeribacter; Celeribacter baekdonensis
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
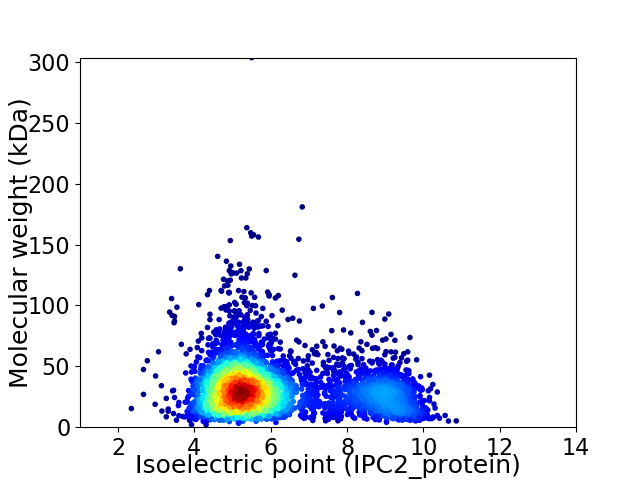
Virtual 2D-PAGE plot for 4198 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K2JW45|K2JW45_9RHOB Protoporphyrinogen IX oxidase OS=Celeribacter baekdonensis B30 OX=1208323 GN=B30_02380 PE=3 SV=1
MM1 pKa = 6.64TNYY4 pKa = 10.58YY5 pKa = 9.48IVSSNPEE12 pKa = 3.57VHH14 pKa = 7.96DD15 pKa = 4.81DD16 pKa = 4.05GNVNNYY22 pKa = 7.25TQVQAGALTPQEE34 pKa = 3.55GDD36 pKa = 3.0AYY38 pKa = 11.03YY39 pKa = 11.27LDD41 pKa = 5.06GNLSSSLWFYY51 pKa = 11.32GSASFTVNIEE61 pKa = 3.82QSLVTNNLVGGATMWIQGTTNATVNIGAGVDD92 pKa = 3.32ADD94 pKa = 3.98YY95 pKa = 11.8LNISSSGDD103 pKa = 3.19TLTINVDD110 pKa = 3.49DD111 pKa = 4.88GATLGAITAPLTDD124 pKa = 3.11QMTIVAGNDD133 pKa = 2.95VTMNGALIGSNGDD146 pKa = 3.41STVTFGSGATFNSTVNFDD164 pKa = 3.09GWGTLSEE171 pKa = 4.41TGSQTFSVGDD181 pKa = 3.53NAVFEE186 pKa = 4.97YY187 pKa = 10.2GVSMDD192 pKa = 3.76GSYY195 pKa = 8.45THH197 pKa = 6.86NSFSAGVNSTIGDD210 pKa = 4.31DD211 pKa = 3.18IVMNGSGYY219 pKa = 10.39INLTDD224 pKa = 3.74DD225 pKa = 3.28TSGYY229 pKa = 9.46SGSVTVGDD237 pKa = 4.04GSTVQGNVYY246 pKa = 9.92MGGSYY251 pKa = 11.49GDD253 pKa = 3.31MTVAVGNDD261 pKa = 3.26VSVGDD266 pKa = 3.88IAVGGSFNDD275 pKa = 3.03IDD277 pKa = 3.67ITVGDD282 pKa = 4.36RR283 pKa = 11.84VDD285 pKa = 2.93TGYY288 pKa = 10.6IGMGGSDD295 pKa = 3.13INGRR299 pKa = 11.84ILVGSEE305 pKa = 4.06GNVSAIYY312 pKa = 10.44AGGSSSDD319 pKa = 3.0ICVGVGDD326 pKa = 4.36NTNIDD331 pKa = 3.58LSIAMGGSNDD341 pKa = 3.73SYY343 pKa = 11.66ALSLGDD349 pKa = 3.78GVSIGLDD356 pKa = 3.19LSMGGSNNVQNVVMGDD372 pKa = 3.66DD373 pKa = 3.87VSVGTYY379 pKa = 10.28VFMGGVDD386 pKa = 3.53NTNHH390 pKa = 5.75VEE392 pKa = 3.64IGNNFTLGSHH402 pKa = 4.9WTGSISGSDD411 pKa = 3.51YY412 pKa = 11.66LEE414 pKa = 5.8IGDD417 pKa = 4.09NWDD420 pKa = 3.39IGTGIYY426 pKa = 10.08LQGGDD431 pKa = 3.9DD432 pKa = 3.9TLILGTVADD441 pKa = 4.85GVTAYY446 pKa = 9.78IDD448 pKa = 3.81GGLGTDD454 pKa = 3.3ALSLRR459 pKa = 11.84VAAGDD464 pKa = 3.52TGFVSAATEE473 pKa = 4.16AGWTYY478 pKa = 11.55DD479 pKa = 3.28SDD481 pKa = 3.72TGTWSSNGNDD491 pKa = 3.2LTYY494 pKa = 11.28NGVTYY499 pKa = 10.41TSFEE503 pKa = 3.96TAAVVDD509 pKa = 4.33EE510 pKa = 4.71DD511 pKa = 4.12WEE513 pKa = 4.61LVCPVANNIVDD524 pKa = 4.09GTAGDD529 pKa = 4.03DD530 pKa = 3.85VMDD533 pKa = 4.46IGYY536 pKa = 8.6TDD538 pKa = 3.6AQGDD542 pKa = 4.13QIDD545 pKa = 4.4GTDD548 pKa = 3.54GDD550 pKa = 3.98TDD552 pKa = 4.19VIFGYY557 pKa = 10.87AGDD560 pKa = 3.82DD561 pKa = 4.1TINGGAGNDD570 pKa = 4.18TIHH573 pKa = 7.12GGLDD577 pKa = 3.18NDD579 pKa = 5.21TITGGAGDD587 pKa = 4.4DD588 pKa = 4.2SVTGGAGSDD597 pKa = 3.49TLAGGVGADD606 pKa = 3.79VLDD609 pKa = 4.68GGSGDD614 pKa = 4.31DD615 pKa = 4.22SLDD618 pKa = 3.52GGEE621 pKa = 5.42GDD623 pKa = 5.57DD624 pKa = 4.49EE625 pKa = 4.84LLGGLGADD633 pKa = 3.91TLIGGAGADD642 pKa = 3.84TLTGGDD648 pKa = 3.76GADD651 pKa = 4.11LFIVDD656 pKa = 5.31DD657 pKa = 4.81GGDD660 pKa = 3.69TITDD664 pKa = 4.14FDD666 pKa = 4.06ATSGIGDD673 pKa = 4.73DD674 pKa = 5.11DD675 pKa = 4.82EE676 pKa = 6.91SDD678 pKa = 3.59NDD680 pKa = 3.94VVDD683 pKa = 5.26LSTFYY688 pKa = 11.48NEE690 pKa = 3.75TTLAEE695 pKa = 4.02WNAANPTQTYY705 pKa = 10.14ASPLGWLRR713 pKa = 11.84ADD715 pKa = 3.53QGDD718 pKa = 4.12GVLDD722 pKa = 3.85AASGLEE728 pKa = 4.32LYY730 pKa = 10.83SPDD733 pKa = 3.63GNPVTSADD741 pKa = 3.69LNAEE745 pKa = 4.06NTRR748 pKa = 11.84VICFARR754 pKa = 11.84GTRR757 pKa = 11.84IMTPRR762 pKa = 11.84GEE764 pKa = 4.62VEE766 pKa = 3.79IQNLKK771 pKa = 10.67VGDD774 pKa = 3.96NVLTLDD780 pKa = 4.45NGFKK784 pKa = 9.46PIRR787 pKa = 11.84WIGHH791 pKa = 5.4RR792 pKa = 11.84RR793 pKa = 11.84LGAEE797 pKa = 4.92ALDD800 pKa = 4.07AMPKK804 pKa = 9.89LRR806 pKa = 11.84PIRR809 pKa = 11.84IRR811 pKa = 11.84AGALDD816 pKa = 3.55TDD818 pKa = 4.17LPVRR822 pKa = 11.84DD823 pKa = 4.24LVVSPQHH830 pKa = 6.14RR831 pKa = 11.84VLFRR835 pKa = 11.84SPHH838 pKa = 5.63AEE840 pKa = 3.85LMFGTHH846 pKa = 5.62EE847 pKa = 4.16VLLAARR853 pKa = 11.84QLLGLEE859 pKa = 4.75GVCVEE864 pKa = 4.45RR865 pKa = 11.84EE866 pKa = 4.09ARR868 pKa = 11.84EE869 pKa = 3.72VEE871 pKa = 4.15YY872 pKa = 10.08WHH874 pKa = 6.74FLFNRR879 pKa = 11.84HH880 pKa = 5.98EE881 pKa = 4.09IVFAEE886 pKa = 5.04GAASEE891 pKa = 4.19SLYY894 pKa = 10.92TGVEE898 pKa = 4.03ALKK901 pKa = 10.77SLNPEE906 pKa = 3.33ARR908 pKa = 11.84EE909 pKa = 3.91EE910 pKa = 4.0IFALFPEE917 pKa = 5.14LGADD921 pKa = 3.39EE922 pKa = 4.52ATRR925 pKa = 11.84KK926 pKa = 9.46PDD928 pKa = 3.34LARR931 pKa = 11.84PVVNGVVARR940 pKa = 11.84LLVSVNGSGQTANILHH956 pKa = 6.7
MM1 pKa = 6.64TNYY4 pKa = 10.58YY5 pKa = 9.48IVSSNPEE12 pKa = 3.57VHH14 pKa = 7.96DD15 pKa = 4.81DD16 pKa = 4.05GNVNNYY22 pKa = 7.25TQVQAGALTPQEE34 pKa = 3.55GDD36 pKa = 3.0AYY38 pKa = 11.03YY39 pKa = 11.27LDD41 pKa = 5.06GNLSSSLWFYY51 pKa = 11.32GSASFTVNIEE61 pKa = 3.82QSLVTNNLVGGATMWIQGTTNATVNIGAGVDD92 pKa = 3.32ADD94 pKa = 3.98YY95 pKa = 11.8LNISSSGDD103 pKa = 3.19TLTINVDD110 pKa = 3.49DD111 pKa = 4.88GATLGAITAPLTDD124 pKa = 3.11QMTIVAGNDD133 pKa = 2.95VTMNGALIGSNGDD146 pKa = 3.41STVTFGSGATFNSTVNFDD164 pKa = 3.09GWGTLSEE171 pKa = 4.41TGSQTFSVGDD181 pKa = 3.53NAVFEE186 pKa = 4.97YY187 pKa = 10.2GVSMDD192 pKa = 3.76GSYY195 pKa = 8.45THH197 pKa = 6.86NSFSAGVNSTIGDD210 pKa = 4.31DD211 pKa = 3.18IVMNGSGYY219 pKa = 10.39INLTDD224 pKa = 3.74DD225 pKa = 3.28TSGYY229 pKa = 9.46SGSVTVGDD237 pKa = 4.04GSTVQGNVYY246 pKa = 9.92MGGSYY251 pKa = 11.49GDD253 pKa = 3.31MTVAVGNDD261 pKa = 3.26VSVGDD266 pKa = 3.88IAVGGSFNDD275 pKa = 3.03IDD277 pKa = 3.67ITVGDD282 pKa = 4.36RR283 pKa = 11.84VDD285 pKa = 2.93TGYY288 pKa = 10.6IGMGGSDD295 pKa = 3.13INGRR299 pKa = 11.84ILVGSEE305 pKa = 4.06GNVSAIYY312 pKa = 10.44AGGSSSDD319 pKa = 3.0ICVGVGDD326 pKa = 4.36NTNIDD331 pKa = 3.58LSIAMGGSNDD341 pKa = 3.73SYY343 pKa = 11.66ALSLGDD349 pKa = 3.78GVSIGLDD356 pKa = 3.19LSMGGSNNVQNVVMGDD372 pKa = 3.66DD373 pKa = 3.87VSVGTYY379 pKa = 10.28VFMGGVDD386 pKa = 3.53NTNHH390 pKa = 5.75VEE392 pKa = 3.64IGNNFTLGSHH402 pKa = 4.9WTGSISGSDD411 pKa = 3.51YY412 pKa = 11.66LEE414 pKa = 5.8IGDD417 pKa = 4.09NWDD420 pKa = 3.39IGTGIYY426 pKa = 10.08LQGGDD431 pKa = 3.9DD432 pKa = 3.9TLILGTVADD441 pKa = 4.85GVTAYY446 pKa = 9.78IDD448 pKa = 3.81GGLGTDD454 pKa = 3.3ALSLRR459 pKa = 11.84VAAGDD464 pKa = 3.52TGFVSAATEE473 pKa = 4.16AGWTYY478 pKa = 11.55DD479 pKa = 3.28SDD481 pKa = 3.72TGTWSSNGNDD491 pKa = 3.2LTYY494 pKa = 11.28NGVTYY499 pKa = 10.41TSFEE503 pKa = 3.96TAAVVDD509 pKa = 4.33EE510 pKa = 4.71DD511 pKa = 4.12WEE513 pKa = 4.61LVCPVANNIVDD524 pKa = 4.09GTAGDD529 pKa = 4.03DD530 pKa = 3.85VMDD533 pKa = 4.46IGYY536 pKa = 8.6TDD538 pKa = 3.6AQGDD542 pKa = 4.13QIDD545 pKa = 4.4GTDD548 pKa = 3.54GDD550 pKa = 3.98TDD552 pKa = 4.19VIFGYY557 pKa = 10.87AGDD560 pKa = 3.82DD561 pKa = 4.1TINGGAGNDD570 pKa = 4.18TIHH573 pKa = 7.12GGLDD577 pKa = 3.18NDD579 pKa = 5.21TITGGAGDD587 pKa = 4.4DD588 pKa = 4.2SVTGGAGSDD597 pKa = 3.49TLAGGVGADD606 pKa = 3.79VLDD609 pKa = 4.68GGSGDD614 pKa = 4.31DD615 pKa = 4.22SLDD618 pKa = 3.52GGEE621 pKa = 5.42GDD623 pKa = 5.57DD624 pKa = 4.49EE625 pKa = 4.84LLGGLGADD633 pKa = 3.91TLIGGAGADD642 pKa = 3.84TLTGGDD648 pKa = 3.76GADD651 pKa = 4.11LFIVDD656 pKa = 5.31DD657 pKa = 4.81GGDD660 pKa = 3.69TITDD664 pKa = 4.14FDD666 pKa = 4.06ATSGIGDD673 pKa = 4.73DD674 pKa = 5.11DD675 pKa = 4.82EE676 pKa = 6.91SDD678 pKa = 3.59NDD680 pKa = 3.94VVDD683 pKa = 5.26LSTFYY688 pKa = 11.48NEE690 pKa = 3.75TTLAEE695 pKa = 4.02WNAANPTQTYY705 pKa = 10.14ASPLGWLRR713 pKa = 11.84ADD715 pKa = 3.53QGDD718 pKa = 4.12GVLDD722 pKa = 3.85AASGLEE728 pKa = 4.32LYY730 pKa = 10.83SPDD733 pKa = 3.63GNPVTSADD741 pKa = 3.69LNAEE745 pKa = 4.06NTRR748 pKa = 11.84VICFARR754 pKa = 11.84GTRR757 pKa = 11.84IMTPRR762 pKa = 11.84GEE764 pKa = 4.62VEE766 pKa = 3.79IQNLKK771 pKa = 10.67VGDD774 pKa = 3.96NVLTLDD780 pKa = 4.45NGFKK784 pKa = 9.46PIRR787 pKa = 11.84WIGHH791 pKa = 5.4RR792 pKa = 11.84RR793 pKa = 11.84LGAEE797 pKa = 4.92ALDD800 pKa = 4.07AMPKK804 pKa = 9.89LRR806 pKa = 11.84PIRR809 pKa = 11.84IRR811 pKa = 11.84AGALDD816 pKa = 3.55TDD818 pKa = 4.17LPVRR822 pKa = 11.84DD823 pKa = 4.24LVVSPQHH830 pKa = 6.14RR831 pKa = 11.84VLFRR835 pKa = 11.84SPHH838 pKa = 5.63AEE840 pKa = 3.85LMFGTHH846 pKa = 5.62EE847 pKa = 4.16VLLAARR853 pKa = 11.84QLLGLEE859 pKa = 4.75GVCVEE864 pKa = 4.45RR865 pKa = 11.84EE866 pKa = 4.09ARR868 pKa = 11.84EE869 pKa = 3.72VEE871 pKa = 4.15YY872 pKa = 10.08WHH874 pKa = 6.74FLFNRR879 pKa = 11.84HH880 pKa = 5.98EE881 pKa = 4.09IVFAEE886 pKa = 5.04GAASEE891 pKa = 4.19SLYY894 pKa = 10.92TGVEE898 pKa = 4.03ALKK901 pKa = 10.77SLNPEE906 pKa = 3.33ARR908 pKa = 11.84EE909 pKa = 3.91EE910 pKa = 4.0IFALFPEE917 pKa = 5.14LGADD921 pKa = 3.39EE922 pKa = 4.52ATRR925 pKa = 11.84KK926 pKa = 9.46PDD928 pKa = 3.34LARR931 pKa = 11.84PVVNGVVARR940 pKa = 11.84LLVSVNGSGQTANILHH956 pKa = 6.7
Molecular weight: 98.55 kDa
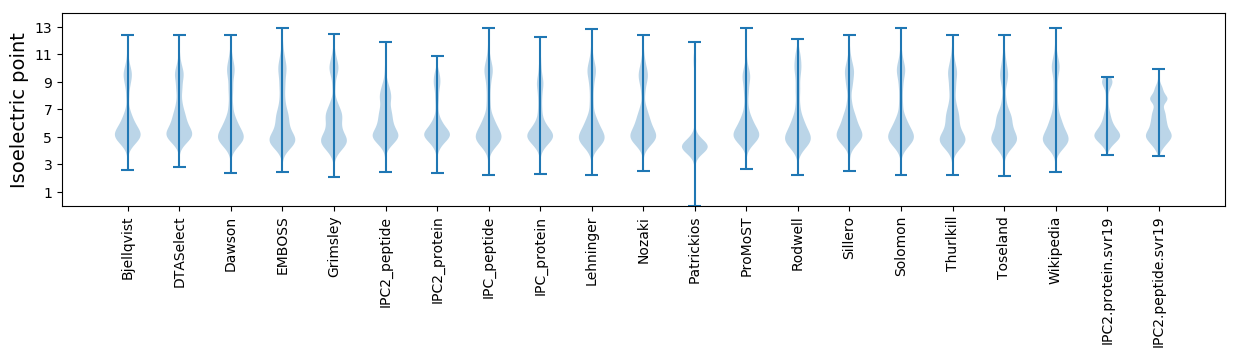
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K2JWU2|K2JWU2_9RHOB Uncharacterized protein OS=Celeribacter baekdonensis B30 OX=1208323 GN=B30_03430 PE=4 SV=1
MM1 pKa = 7.53KK2 pKa = 7.9PTPRR6 pKa = 11.84DD7 pKa = 3.25LVITRR12 pKa = 11.84WGARR16 pKa = 11.84FMGRR20 pKa = 11.84RR21 pKa = 11.84FPCSIGKK28 pKa = 10.06GGIVANKK35 pKa = 9.98RR36 pKa = 11.84EE37 pKa = 4.36GDD39 pKa = 3.41GGTPIGPHH47 pKa = 6.85EE48 pKa = 4.25IAEE51 pKa = 4.3LYY53 pKa = 10.12YY54 pKa = 10.78RR55 pKa = 11.84ADD57 pKa = 3.69RR58 pKa = 11.84VPALNGTEE66 pKa = 3.82IRR68 pKa = 11.84IGDD71 pKa = 4.58LWCDD75 pKa = 3.37ATDD78 pKa = 3.51HH79 pKa = 6.53PRR81 pKa = 11.84YY82 pKa = 9.11NHH84 pKa = 6.91LVRR87 pKa = 11.84APFKK91 pKa = 10.69ASHH94 pKa = 5.66EE95 pKa = 4.14KK96 pKa = 9.25MRR98 pKa = 11.84RR99 pKa = 11.84GDD101 pKa = 3.38RR102 pKa = 11.84LYY104 pKa = 11.38DD105 pKa = 4.11LVLTTNWNWPEE116 pKa = 3.75ATPGKK121 pKa = 10.34GSAIFIHH128 pKa = 6.76RR129 pKa = 11.84WRR131 pKa = 11.84KK132 pKa = 8.14PRR134 pKa = 11.84HH135 pKa = 4.52PTEE138 pKa = 4.0GCVAFRR144 pKa = 11.84ADD146 pKa = 3.42HH147 pKa = 6.11LRR149 pKa = 11.84WIAEE153 pKa = 4.3RR154 pKa = 11.84ISPATRR160 pKa = 11.84LIVRR164 pKa = 4.59
MM1 pKa = 7.53KK2 pKa = 7.9PTPRR6 pKa = 11.84DD7 pKa = 3.25LVITRR12 pKa = 11.84WGARR16 pKa = 11.84FMGRR20 pKa = 11.84RR21 pKa = 11.84FPCSIGKK28 pKa = 10.06GGIVANKK35 pKa = 9.98RR36 pKa = 11.84EE37 pKa = 4.36GDD39 pKa = 3.41GGTPIGPHH47 pKa = 6.85EE48 pKa = 4.25IAEE51 pKa = 4.3LYY53 pKa = 10.12YY54 pKa = 10.78RR55 pKa = 11.84ADD57 pKa = 3.69RR58 pKa = 11.84VPALNGTEE66 pKa = 3.82IRR68 pKa = 11.84IGDD71 pKa = 4.58LWCDD75 pKa = 3.37ATDD78 pKa = 3.51HH79 pKa = 6.53PRR81 pKa = 11.84YY82 pKa = 9.11NHH84 pKa = 6.91LVRR87 pKa = 11.84APFKK91 pKa = 10.69ASHH94 pKa = 5.66EE95 pKa = 4.14KK96 pKa = 9.25MRR98 pKa = 11.84RR99 pKa = 11.84GDD101 pKa = 3.38RR102 pKa = 11.84LYY104 pKa = 11.38DD105 pKa = 4.11LVLTTNWNWPEE116 pKa = 3.75ATPGKK121 pKa = 10.34GSAIFIHH128 pKa = 6.76RR129 pKa = 11.84WRR131 pKa = 11.84KK132 pKa = 8.14PRR134 pKa = 11.84HH135 pKa = 4.52PTEE138 pKa = 4.0GCVAFRR144 pKa = 11.84ADD146 pKa = 3.42HH147 pKa = 6.11LRR149 pKa = 11.84WIAEE153 pKa = 4.3RR154 pKa = 11.84ISPATRR160 pKa = 11.84LIVRR164 pKa = 4.59
Molecular weight: 18.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1288073 |
19 |
2756 |
306.8 |
33.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.833 ± 0.05 | 0.891 ± 0.012 |
6.009 ± 0.032 | 5.829 ± 0.038 |
3.867 ± 0.023 | 8.489 ± 0.039 |
2.118 ± 0.017 | 5.431 ± 0.025 |
3.503 ± 0.029 | 10.07 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.921 ± 0.016 | 2.615 ± 0.021 |
4.962 ± 0.027 | 3.119 ± 0.02 |
6.238 ± 0.037 | 5.437 ± 0.026 |
5.742 ± 0.028 | 7.312 ± 0.031 |
1.334 ± 0.017 | 2.28 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
