
Cestrum yellow leaf curling virus (CmYLCV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Soymovirus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
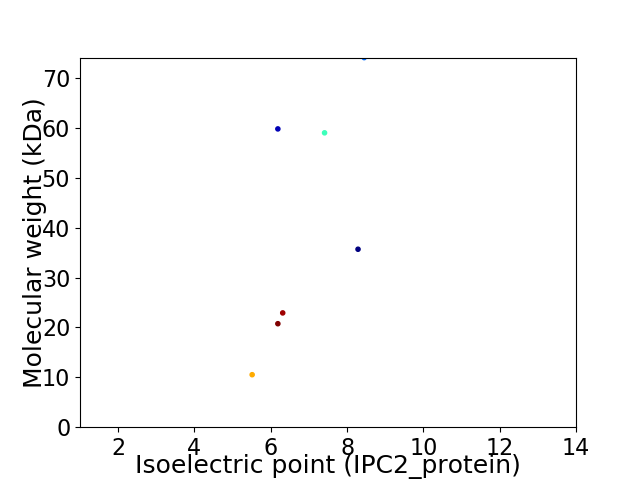
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
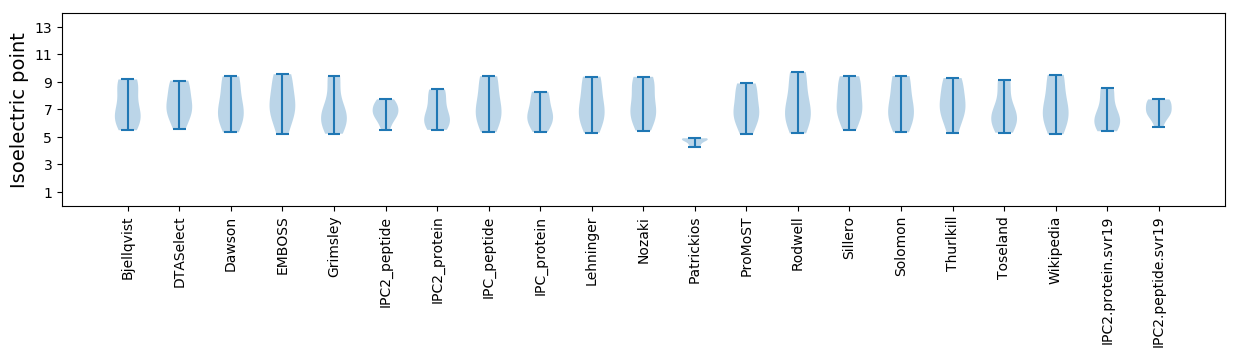
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q7TD13|MVP_CYLCV Putative movement protein OS=Cestrum yellow leaf curling virus OX=175814 GN=ORF I PE=3 SV=1
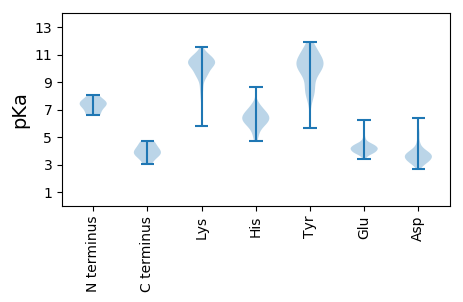
MM1 pKa = 6.71SQEE4 pKa = 4.25RR5 pKa = 11.84VDD7 pKa = 3.7KK8 pKa = 10.8LRR10 pKa = 11.84KK11 pKa = 9.51LRR13 pKa = 11.84IEE15 pKa = 4.01FEE17 pKa = 4.48GIIQRR22 pKa = 11.84MEE24 pKa = 3.75VAKK27 pKa = 10.84NLLRR31 pKa = 11.84EE32 pKa = 4.35LEE34 pKa = 4.19EE35 pKa = 3.88TTDD38 pKa = 3.25RR39 pKa = 11.84VRR41 pKa = 11.84VDD43 pKa = 3.51RR44 pKa = 11.84LQMEE48 pKa = 4.5VGFLRR53 pKa = 11.84DD54 pKa = 3.5YY55 pKa = 11.94ADD57 pKa = 3.63INCMIEE63 pKa = 3.8KK64 pKa = 10.47DD65 pKa = 3.75RR66 pKa = 11.84IKK68 pKa = 10.28EE69 pKa = 3.95QSSSSSATRR78 pKa = 11.84TTQEE82 pKa = 4.2PSLHH86 pKa = 6.6LPDD89 pKa = 3.98
MM1 pKa = 6.71SQEE4 pKa = 4.25RR5 pKa = 11.84VDD7 pKa = 3.7KK8 pKa = 10.8LRR10 pKa = 11.84KK11 pKa = 9.51LRR13 pKa = 11.84IEE15 pKa = 4.01FEE17 pKa = 4.48GIIQRR22 pKa = 11.84MEE24 pKa = 3.75VAKK27 pKa = 10.84NLLRR31 pKa = 11.84EE32 pKa = 4.35LEE34 pKa = 4.19EE35 pKa = 3.88TTDD38 pKa = 3.25RR39 pKa = 11.84VRR41 pKa = 11.84VDD43 pKa = 3.51RR44 pKa = 11.84LQMEE48 pKa = 4.5VGFLRR53 pKa = 11.84DD54 pKa = 3.5YY55 pKa = 11.94ADD57 pKa = 3.63INCMIEE63 pKa = 3.8KK64 pKa = 10.47DD65 pKa = 3.75RR66 pKa = 11.84IKK68 pKa = 10.28EE69 pKa = 3.95QSSSSSATRR78 pKa = 11.84TTQEE82 pKa = 4.2PSLHH86 pKa = 6.6LPDD89 pKa = 3.98
Molecular weight: 10.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q7TD09|CAPSD_CYLCV Capsid protein OS=Cestrum yellow leaf curling virus OX=175814 GN=ORF IV PE=3 SV=1
MM1 pKa = 7.51KK2 pKa = 10.69SKK4 pKa = 10.84GNPNATFITVKK15 pKa = 10.51INDD18 pKa = 3.78VFINAYY24 pKa = 9.51VDD26 pKa = 3.57TGATICLADD35 pKa = 4.29PKK37 pKa = 10.8IKK39 pKa = 10.7LKK41 pKa = 8.32WVKK44 pKa = 8.92MEE46 pKa = 4.13KK47 pKa = 9.85PIKK50 pKa = 10.15ISIADD55 pKa = 3.65KK56 pKa = 9.91SVQEE60 pKa = 3.63IWHH63 pKa = 5.86RR64 pKa = 11.84AEE66 pKa = 4.6MVEE69 pKa = 3.63IWIRR73 pKa = 11.84NYY75 pKa = 10.62KK76 pKa = 9.68FVAATVCQKK85 pKa = 10.91SSGMDD90 pKa = 3.02FVIGNNFLRR99 pKa = 11.84LYY101 pKa = 10.62QPFIQGLNYY110 pKa = 9.78IKK112 pKa = 10.66LRR114 pKa = 11.84APLDD118 pKa = 3.44KK119 pKa = 10.91DD120 pKa = 3.18INQPSKK126 pKa = 10.34MIYY129 pKa = 9.97IPVTTPSKK137 pKa = 10.18ILQFAILEE145 pKa = 4.15KK146 pKa = 10.78LQDD149 pKa = 3.57ILFEE153 pKa = 4.12LHH155 pKa = 5.48VQEE158 pKa = 4.74NSKK161 pKa = 9.08TPLEE165 pKa = 4.38LKK167 pKa = 10.58VSSTLEE173 pKa = 4.09EE174 pKa = 4.27VCDD177 pKa = 4.13EE178 pKa = 4.5NPLDD182 pKa = 3.88VKK184 pKa = 10.04NTNTEE189 pKa = 3.87LVKK192 pKa = 10.44IEE194 pKa = 4.91LINPEE199 pKa = 4.19KK200 pKa = 10.46EE201 pKa = 4.11VNVPNNIPYY210 pKa = 10.29SLRR213 pKa = 11.84DD214 pKa = 3.34INEE217 pKa = 4.23FSQEE221 pKa = 3.98CADD224 pKa = 4.17LVRR227 pKa = 11.84KK228 pKa = 9.91GIIEE232 pKa = 4.08EE233 pKa = 4.41SKK235 pKa = 10.75SPHH238 pKa = 6.11SAPAFYY244 pKa = 10.93VEE246 pKa = 3.96NHH248 pKa = 5.87NEE250 pKa = 3.65IKK252 pKa = 10.1RR253 pKa = 11.84KK254 pKa = 8.33KK255 pKa = 8.87RR256 pKa = 11.84RR257 pKa = 11.84MVINYY262 pKa = 7.65KK263 pKa = 10.32ALNKK267 pKa = 9.17ATIGNAHH274 pKa = 7.33KK275 pKa = 10.52LPRR278 pKa = 11.84IDD280 pKa = 5.46SILTKK285 pKa = 10.75VKK287 pKa = 10.01GSNWFSTLDD296 pKa = 3.37AKK298 pKa = 10.57SGYY301 pKa = 7.83WQLRR305 pKa = 11.84LHH307 pKa = 6.32PQSKK311 pKa = 9.67PLTAFSCPPQKK322 pKa = 10.42HH323 pKa = 4.95YY324 pKa = 10.35QWNVLPFGLKK334 pKa = 8.94QAPGIYY340 pKa = 9.8QNFMDD345 pKa = 5.07KK346 pKa = 10.78NLEE349 pKa = 3.97GLEE352 pKa = 4.29NFCLAYY358 pKa = 9.76IDD360 pKa = 5.6DD361 pKa = 4.18ILVFTNSSRR370 pKa = 11.84EE371 pKa = 3.86EE372 pKa = 4.13HH373 pKa = 6.21LSKK376 pKa = 10.98LLVVLEE382 pKa = 4.09RR383 pKa = 11.84CKK385 pKa = 10.93EE386 pKa = 3.8KK387 pKa = 11.11GLILSKK393 pKa = 10.56KK394 pKa = 8.55KK395 pKa = 10.74AIIARR400 pKa = 11.84QTIDD404 pKa = 3.73FLGLTLQEE412 pKa = 4.1NGEE415 pKa = 4.31IKK417 pKa = 10.47LQPNVLEE424 pKa = 4.28KK425 pKa = 11.42LEE427 pKa = 4.6LFPDD431 pKa = 4.64AIEE434 pKa = 5.46DD435 pKa = 3.63RR436 pKa = 11.84KK437 pKa = 10.26QLQRR441 pKa = 11.84FLGCLNYY448 pKa = 9.98IADD451 pKa = 3.78KK452 pKa = 11.38GFLKK456 pKa = 10.68EE457 pKa = 3.8IAKK460 pKa = 7.49EE461 pKa = 4.3TKK463 pKa = 9.51NLYY466 pKa = 9.85PKK468 pKa = 10.78VSITNPWHH476 pKa = 6.39WSDD479 pKa = 5.14LDD481 pKa = 3.93SKK483 pKa = 11.07LVNQIKK489 pKa = 10.27KK490 pKa = 9.43KK491 pKa = 10.83CKK493 pKa = 9.91DD494 pKa = 3.46LSPLYY499 pKa = 10.05FPKK502 pKa = 10.71PEE504 pKa = 5.05DD505 pKa = 3.52YY506 pKa = 11.22LIIEE510 pKa = 4.65TDD512 pKa = 3.18ASGDD516 pKa = 3.26TWAGCLKK523 pKa = 10.42AAEE526 pKa = 4.35LLFPKK531 pKa = 9.43GTKK534 pKa = 9.7NKK536 pKa = 9.81VVEE539 pKa = 4.38RR540 pKa = 11.84LCKK543 pKa = 7.76YY544 pKa = 8.67TSGIFSSAEE553 pKa = 3.51QKK555 pKa = 9.33YY556 pKa = 7.05TVHH559 pKa = 6.39EE560 pKa = 4.52KK561 pKa = 9.01EE562 pKa = 4.01TLAALKK568 pKa = 9.71TMRR571 pKa = 11.84KK572 pKa = 7.89WKK574 pKa = 10.82AEE576 pKa = 3.98LLPKK580 pKa = 10.42EE581 pKa = 4.03FTLRR585 pKa = 11.84TDD587 pKa = 2.87SSYY590 pKa = 8.73VTGFARR596 pKa = 11.84HH597 pKa = 5.41NLKK600 pKa = 10.81ANYY603 pKa = 9.1NQGRR607 pKa = 11.84LVRR610 pKa = 11.84WQLEE614 pKa = 3.95FLQYY618 pKa = 9.13PARR621 pKa = 11.84VEE623 pKa = 4.16YY624 pKa = 10.46IKK626 pKa = 11.13GEE628 pKa = 4.27KK629 pKa = 10.01NSLADD634 pKa = 3.34TLTRR638 pKa = 11.84EE639 pKa = 4.59WKK641 pKa = 9.22QQQ643 pKa = 3.03
MM1 pKa = 7.51KK2 pKa = 10.69SKK4 pKa = 10.84GNPNATFITVKK15 pKa = 10.51INDD18 pKa = 3.78VFINAYY24 pKa = 9.51VDD26 pKa = 3.57TGATICLADD35 pKa = 4.29PKK37 pKa = 10.8IKK39 pKa = 10.7LKK41 pKa = 8.32WVKK44 pKa = 8.92MEE46 pKa = 4.13KK47 pKa = 9.85PIKK50 pKa = 10.15ISIADD55 pKa = 3.65KK56 pKa = 9.91SVQEE60 pKa = 3.63IWHH63 pKa = 5.86RR64 pKa = 11.84AEE66 pKa = 4.6MVEE69 pKa = 3.63IWIRR73 pKa = 11.84NYY75 pKa = 10.62KK76 pKa = 9.68FVAATVCQKK85 pKa = 10.91SSGMDD90 pKa = 3.02FVIGNNFLRR99 pKa = 11.84LYY101 pKa = 10.62QPFIQGLNYY110 pKa = 9.78IKK112 pKa = 10.66LRR114 pKa = 11.84APLDD118 pKa = 3.44KK119 pKa = 10.91DD120 pKa = 3.18INQPSKK126 pKa = 10.34MIYY129 pKa = 9.97IPVTTPSKK137 pKa = 10.18ILQFAILEE145 pKa = 4.15KK146 pKa = 10.78LQDD149 pKa = 3.57ILFEE153 pKa = 4.12LHH155 pKa = 5.48VQEE158 pKa = 4.74NSKK161 pKa = 9.08TPLEE165 pKa = 4.38LKK167 pKa = 10.58VSSTLEE173 pKa = 4.09EE174 pKa = 4.27VCDD177 pKa = 4.13EE178 pKa = 4.5NPLDD182 pKa = 3.88VKK184 pKa = 10.04NTNTEE189 pKa = 3.87LVKK192 pKa = 10.44IEE194 pKa = 4.91LINPEE199 pKa = 4.19KK200 pKa = 10.46EE201 pKa = 4.11VNVPNNIPYY210 pKa = 10.29SLRR213 pKa = 11.84DD214 pKa = 3.34INEE217 pKa = 4.23FSQEE221 pKa = 3.98CADD224 pKa = 4.17LVRR227 pKa = 11.84KK228 pKa = 9.91GIIEE232 pKa = 4.08EE233 pKa = 4.41SKK235 pKa = 10.75SPHH238 pKa = 6.11SAPAFYY244 pKa = 10.93VEE246 pKa = 3.96NHH248 pKa = 5.87NEE250 pKa = 3.65IKK252 pKa = 10.1RR253 pKa = 11.84KK254 pKa = 8.33KK255 pKa = 8.87RR256 pKa = 11.84RR257 pKa = 11.84MVINYY262 pKa = 7.65KK263 pKa = 10.32ALNKK267 pKa = 9.17ATIGNAHH274 pKa = 7.33KK275 pKa = 10.52LPRR278 pKa = 11.84IDD280 pKa = 5.46SILTKK285 pKa = 10.75VKK287 pKa = 10.01GSNWFSTLDD296 pKa = 3.37AKK298 pKa = 10.57SGYY301 pKa = 7.83WQLRR305 pKa = 11.84LHH307 pKa = 6.32PQSKK311 pKa = 9.67PLTAFSCPPQKK322 pKa = 10.42HH323 pKa = 4.95YY324 pKa = 10.35QWNVLPFGLKK334 pKa = 8.94QAPGIYY340 pKa = 9.8QNFMDD345 pKa = 5.07KK346 pKa = 10.78NLEE349 pKa = 3.97GLEE352 pKa = 4.29NFCLAYY358 pKa = 9.76IDD360 pKa = 5.6DD361 pKa = 4.18ILVFTNSSRR370 pKa = 11.84EE371 pKa = 3.86EE372 pKa = 4.13HH373 pKa = 6.21LSKK376 pKa = 10.98LLVVLEE382 pKa = 4.09RR383 pKa = 11.84CKK385 pKa = 10.93EE386 pKa = 3.8KK387 pKa = 11.11GLILSKK393 pKa = 10.56KK394 pKa = 8.55KK395 pKa = 10.74AIIARR400 pKa = 11.84QTIDD404 pKa = 3.73FLGLTLQEE412 pKa = 4.1NGEE415 pKa = 4.31IKK417 pKa = 10.47LQPNVLEE424 pKa = 4.28KK425 pKa = 11.42LEE427 pKa = 4.6LFPDD431 pKa = 4.64AIEE434 pKa = 5.46DD435 pKa = 3.63RR436 pKa = 11.84KK437 pKa = 10.26QLQRR441 pKa = 11.84FLGCLNYY448 pKa = 9.98IADD451 pKa = 3.78KK452 pKa = 11.38GFLKK456 pKa = 10.68EE457 pKa = 3.8IAKK460 pKa = 7.49EE461 pKa = 4.3TKK463 pKa = 9.51NLYY466 pKa = 9.85PKK468 pKa = 10.78VSITNPWHH476 pKa = 6.39WSDD479 pKa = 5.14LDD481 pKa = 3.93SKK483 pKa = 11.07LVNQIKK489 pKa = 10.27KK490 pKa = 9.43KK491 pKa = 10.83CKK493 pKa = 9.91DD494 pKa = 3.46LSPLYY499 pKa = 10.05FPKK502 pKa = 10.71PEE504 pKa = 5.05DD505 pKa = 3.52YY506 pKa = 11.22LIIEE510 pKa = 4.65TDD512 pKa = 3.18ASGDD516 pKa = 3.26TWAGCLKK523 pKa = 10.42AAEE526 pKa = 4.35LLFPKK531 pKa = 9.43GTKK534 pKa = 9.7NKK536 pKa = 9.81VVEE539 pKa = 4.38RR540 pKa = 11.84LCKK543 pKa = 7.76YY544 pKa = 8.67TSGIFSSAEE553 pKa = 3.51QKK555 pKa = 9.33YY556 pKa = 7.05TVHH559 pKa = 6.39EE560 pKa = 4.52KK561 pKa = 9.01EE562 pKa = 4.01TLAALKK568 pKa = 9.71TMRR571 pKa = 11.84KK572 pKa = 7.89WKK574 pKa = 10.82AEE576 pKa = 3.98LLPKK580 pKa = 10.42EE581 pKa = 4.03FTLRR585 pKa = 11.84TDD587 pKa = 2.87SSYY590 pKa = 8.73VTGFARR596 pKa = 11.84HH597 pKa = 5.41NLKK600 pKa = 10.81ANYY603 pKa = 9.1NQGRR607 pKa = 11.84LVRR610 pKa = 11.84WQLEE614 pKa = 3.95FLQYY618 pKa = 9.13PARR621 pKa = 11.84VEE623 pKa = 4.16YY624 pKa = 10.46IKK626 pKa = 11.13GEE628 pKa = 4.27KK629 pKa = 10.01NSLADD634 pKa = 3.34TLTRR638 pKa = 11.84EE639 pKa = 4.59WKK641 pKa = 9.22QQQ643 pKa = 3.03
Molecular weight: 74.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2455 |
89 |
643 |
350.7 |
40.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.807 ± 0.592 | 1.711 ± 0.34 |
4.888 ± 0.463 | 9.328 ± 0.839 |
4.196 ± 0.29 | 4.033 ± 0.286 |
1.548 ± 0.3 | 7.617 ± 0.411 |
11.039 ± 0.511 | 8.88 ± 1.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.2 ± 0.486 | 5.092 ± 0.515 |
4.684 ± 0.343 | 4.644 ± 0.427 |
4.236 ± 0.561 | 7.291 ± 0.819 |
5.092 ± 0.174 | 4.562 ± 0.586 |
1.018 ± 0.304 | 3.136 ± 0.483 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
