
Sphaeropsis sapinea RNA virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q9YXE7|Q9YXE7_9VIRU Coat protein OS=Sphaeropsis sapinea RNA virus 1 OX=73497 PE=4 SV=1
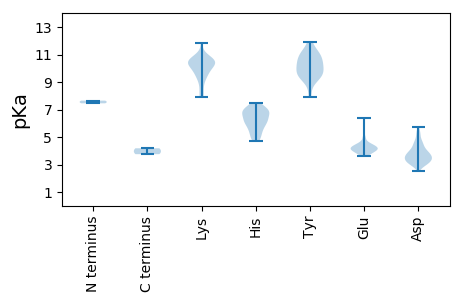
MM1 pKa = 7.65PPISTAQWYY10 pKa = 9.35HH11 pKa = 5.63SLLLKK16 pKa = 10.75SLFVNRR22 pKa = 11.84FRR24 pKa = 11.84QSSLFGGRR32 pKa = 11.84VPLIPNTTIDD42 pKa = 3.95GYY44 pKa = 9.62ITTHH48 pKa = 5.19QHH50 pKa = 4.68HH51 pKa = 6.07TTPNFPMEE59 pKa = 4.19HH60 pKa = 6.88ALRR63 pKa = 11.84NAFLAAVIASPRR75 pKa = 11.84GGTLLTAPTKK85 pKa = 10.3KK86 pKa = 10.44GKK88 pKa = 8.93EE89 pKa = 3.95KK90 pKa = 10.42KK91 pKa = 10.43APLATYY97 pKa = 9.71RR98 pKa = 11.84RR99 pKa = 11.84YY100 pKa = 9.78RR101 pKa = 11.84ANIRR105 pKa = 11.84TEE107 pKa = 3.77STIAGNVDD115 pKa = 2.82ARR117 pKa = 11.84VAGIQYY123 pKa = 9.82EE124 pKa = 3.76IGARR128 pKa = 11.84YY129 pKa = 9.07EE130 pKa = 4.09RR131 pKa = 11.84VGEE134 pKa = 4.02AFAPVPEE141 pKa = 4.13EE142 pKa = 3.66QVLFEE147 pKa = 4.08AAYY150 pKa = 9.16PSSAALAEE158 pKa = 4.36DD159 pKa = 3.91FVGFARR165 pKa = 11.84KK166 pKa = 9.52YY167 pKa = 9.7SNFSATFAHH176 pKa = 6.78SSLAGLVEE184 pKa = 4.46RR185 pKa = 11.84VARR188 pKa = 11.84TLGALTVFPSGTFDD202 pKa = 3.05QDD204 pKa = 3.55AIRR207 pKa = 11.84GGRR210 pKa = 11.84PLMIAALGTLDD221 pKa = 4.52GPVNSLAGSVFIPRR235 pKa = 11.84LVDD238 pKa = 3.7SVISPDD244 pKa = 3.37VFTILINAAAGEE256 pKa = 4.37GSRR259 pKa = 11.84VITDD263 pKa = 3.5VLEE266 pKa = 4.52LDD268 pKa = 3.27ATTRR272 pKa = 11.84RR273 pKa = 11.84PIVPTLRR280 pKa = 11.84DD281 pKa = 3.22SSMLLPCVEE290 pKa = 4.4ALRR293 pKa = 11.84ILGANMAACDD303 pKa = 3.93QGPLFAFALVRR314 pKa = 11.84GLNAVLSLVGHH325 pKa = 5.75TDD327 pKa = 3.14EE328 pKa = 6.4AGVTRR333 pKa = 11.84DD334 pKa = 2.94IFRR337 pKa = 11.84VSGFDD342 pKa = 3.09IPFGGIHH349 pKa = 6.86FGLEE353 pKa = 4.17PYY355 pKa = 10.67AGLPAVASNAGADD368 pKa = 3.41ACCYY372 pKa = 10.64VDD374 pKa = 4.53ALLMTSGALVAHH386 pKa = 7.19CDD388 pKa = 3.33PGQEE392 pKa = 4.04YY393 pKa = 10.38GGRR396 pKa = 11.84WFPTVLQGTGPDD408 pKa = 3.53TAEE411 pKa = 3.82VRR413 pKa = 11.84PGQSQEE419 pKa = 3.79GTADD423 pKa = 3.12MANRR427 pKa = 11.84NRR429 pKa = 11.84GLLASSMPRR438 pKa = 11.84FAEE441 pKa = 4.62HH442 pKa = 6.27YY443 pKa = 10.31ARR445 pKa = 11.84GLGRR449 pKa = 11.84LFAIEE454 pKa = 3.69GDD456 pKa = 3.28ARR458 pKa = 11.84IVVNILSASARR469 pKa = 11.84LLPDD473 pKa = 2.89NCRR476 pKa = 11.84HH477 pKa = 5.59LRR479 pKa = 11.84YY480 pKa = 9.33PSVSPYY486 pKa = 10.22FWVEE490 pKa = 3.85PTSLLPPDD498 pKa = 4.44FLGTAAEE505 pKa = 4.26LNGCGSLAMRR515 pKa = 11.84GTSRR519 pKa = 11.84TRR521 pKa = 11.84QAWEE525 pKa = 4.36DD526 pKa = 3.34IEE528 pKa = 5.0RR529 pKa = 11.84VGDD532 pKa = 3.58EE533 pKa = 4.47DD534 pKa = 4.53VTFTGYY540 pKa = 10.15NVAMPFARR548 pKa = 11.84SSWFLIHH555 pKa = 6.88WLNHH559 pKa = 5.39PANGLGALRR568 pKa = 11.84VRR570 pKa = 11.84QLDD573 pKa = 3.84PNGVIHH579 pKa = 7.1PGPCADD585 pKa = 4.09NPDD588 pKa = 3.12VRR590 pKa = 11.84DD591 pKa = 3.44RR592 pKa = 11.84VEE594 pKa = 4.02ASLPITDD601 pKa = 4.11YY602 pKa = 11.27LWVRR606 pKa = 11.84GQSPFPAPGEE616 pKa = 3.96FLNLTGTLGLLARR629 pKa = 11.84HH630 pKa = 5.93LTMDD634 pKa = 4.71DD635 pKa = 3.69DD636 pKa = 5.37GIPTEE641 pKa = 4.03EE642 pKa = 4.26HH643 pKa = 6.72LPTGRR648 pKa = 11.84EE649 pKa = 3.78FRR651 pKa = 11.84DD652 pKa = 3.37TAVTVVAGRR661 pKa = 11.84PVGLPNGAHH670 pKa = 6.39NAYY673 pKa = 10.26DD674 pKa = 3.66SQARR678 pKa = 11.84RR679 pKa = 11.84ARR681 pKa = 11.84TRR683 pKa = 11.84ATRR686 pKa = 11.84EE687 pKa = 3.73LAASAARR694 pKa = 11.84ARR696 pKa = 11.84LYY698 pKa = 11.07GRR700 pKa = 11.84ATVAEE705 pKa = 4.27MPILTSAPVLRR716 pKa = 11.84AAPPRR721 pKa = 11.84PEE723 pKa = 4.19AVNPMEE729 pKa = 4.98GGGPGHH735 pKa = 7.34DD736 pKa = 4.0LARR739 pKa = 11.84HH740 pKa = 5.42SGMGRR745 pKa = 11.84HH746 pKa = 5.96TYY748 pKa = 10.23PPDD751 pKa = 3.53RR752 pKa = 11.84EE753 pKa = 4.28QRR755 pKa = 11.84GDD757 pKa = 3.43PAVPVPQHH765 pKa = 4.83QALRR769 pKa = 11.84APQMPRR775 pKa = 11.84PGPNPQGGGVIPPPPPLPPSGGGDD799 pKa = 3.33GSGPSSGGSCAQEE812 pKa = 4.32DD813 pKa = 4.8TNPAPPVAPGPAQPDD828 pKa = 4.06GPANEE833 pKa = 4.18
MM1 pKa = 7.65PPISTAQWYY10 pKa = 9.35HH11 pKa = 5.63SLLLKK16 pKa = 10.75SLFVNRR22 pKa = 11.84FRR24 pKa = 11.84QSSLFGGRR32 pKa = 11.84VPLIPNTTIDD42 pKa = 3.95GYY44 pKa = 9.62ITTHH48 pKa = 5.19QHH50 pKa = 4.68HH51 pKa = 6.07TTPNFPMEE59 pKa = 4.19HH60 pKa = 6.88ALRR63 pKa = 11.84NAFLAAVIASPRR75 pKa = 11.84GGTLLTAPTKK85 pKa = 10.3KK86 pKa = 10.44GKK88 pKa = 8.93EE89 pKa = 3.95KK90 pKa = 10.42KK91 pKa = 10.43APLATYY97 pKa = 9.71RR98 pKa = 11.84RR99 pKa = 11.84YY100 pKa = 9.78RR101 pKa = 11.84ANIRR105 pKa = 11.84TEE107 pKa = 3.77STIAGNVDD115 pKa = 2.82ARR117 pKa = 11.84VAGIQYY123 pKa = 9.82EE124 pKa = 3.76IGARR128 pKa = 11.84YY129 pKa = 9.07EE130 pKa = 4.09RR131 pKa = 11.84VGEE134 pKa = 4.02AFAPVPEE141 pKa = 4.13EE142 pKa = 3.66QVLFEE147 pKa = 4.08AAYY150 pKa = 9.16PSSAALAEE158 pKa = 4.36DD159 pKa = 3.91FVGFARR165 pKa = 11.84KK166 pKa = 9.52YY167 pKa = 9.7SNFSATFAHH176 pKa = 6.78SSLAGLVEE184 pKa = 4.46RR185 pKa = 11.84VARR188 pKa = 11.84TLGALTVFPSGTFDD202 pKa = 3.05QDD204 pKa = 3.55AIRR207 pKa = 11.84GGRR210 pKa = 11.84PLMIAALGTLDD221 pKa = 4.52GPVNSLAGSVFIPRR235 pKa = 11.84LVDD238 pKa = 3.7SVISPDD244 pKa = 3.37VFTILINAAAGEE256 pKa = 4.37GSRR259 pKa = 11.84VITDD263 pKa = 3.5VLEE266 pKa = 4.52LDD268 pKa = 3.27ATTRR272 pKa = 11.84RR273 pKa = 11.84PIVPTLRR280 pKa = 11.84DD281 pKa = 3.22SSMLLPCVEE290 pKa = 4.4ALRR293 pKa = 11.84ILGANMAACDD303 pKa = 3.93QGPLFAFALVRR314 pKa = 11.84GLNAVLSLVGHH325 pKa = 5.75TDD327 pKa = 3.14EE328 pKa = 6.4AGVTRR333 pKa = 11.84DD334 pKa = 2.94IFRR337 pKa = 11.84VSGFDD342 pKa = 3.09IPFGGIHH349 pKa = 6.86FGLEE353 pKa = 4.17PYY355 pKa = 10.67AGLPAVASNAGADD368 pKa = 3.41ACCYY372 pKa = 10.64VDD374 pKa = 4.53ALLMTSGALVAHH386 pKa = 7.19CDD388 pKa = 3.33PGQEE392 pKa = 4.04YY393 pKa = 10.38GGRR396 pKa = 11.84WFPTVLQGTGPDD408 pKa = 3.53TAEE411 pKa = 3.82VRR413 pKa = 11.84PGQSQEE419 pKa = 3.79GTADD423 pKa = 3.12MANRR427 pKa = 11.84NRR429 pKa = 11.84GLLASSMPRR438 pKa = 11.84FAEE441 pKa = 4.62HH442 pKa = 6.27YY443 pKa = 10.31ARR445 pKa = 11.84GLGRR449 pKa = 11.84LFAIEE454 pKa = 3.69GDD456 pKa = 3.28ARR458 pKa = 11.84IVVNILSASARR469 pKa = 11.84LLPDD473 pKa = 2.89NCRR476 pKa = 11.84HH477 pKa = 5.59LRR479 pKa = 11.84YY480 pKa = 9.33PSVSPYY486 pKa = 10.22FWVEE490 pKa = 3.85PTSLLPPDD498 pKa = 4.44FLGTAAEE505 pKa = 4.26LNGCGSLAMRR515 pKa = 11.84GTSRR519 pKa = 11.84TRR521 pKa = 11.84QAWEE525 pKa = 4.36DD526 pKa = 3.34IEE528 pKa = 5.0RR529 pKa = 11.84VGDD532 pKa = 3.58EE533 pKa = 4.47DD534 pKa = 4.53VTFTGYY540 pKa = 10.15NVAMPFARR548 pKa = 11.84SSWFLIHH555 pKa = 6.88WLNHH559 pKa = 5.39PANGLGALRR568 pKa = 11.84VRR570 pKa = 11.84QLDD573 pKa = 3.84PNGVIHH579 pKa = 7.1PGPCADD585 pKa = 4.09NPDD588 pKa = 3.12VRR590 pKa = 11.84DD591 pKa = 3.44RR592 pKa = 11.84VEE594 pKa = 4.02ASLPITDD601 pKa = 4.11YY602 pKa = 11.27LWVRR606 pKa = 11.84GQSPFPAPGEE616 pKa = 3.96FLNLTGTLGLLARR629 pKa = 11.84HH630 pKa = 5.93LTMDD634 pKa = 4.71DD635 pKa = 3.69DD636 pKa = 5.37GIPTEE641 pKa = 4.03EE642 pKa = 4.26HH643 pKa = 6.72LPTGRR648 pKa = 11.84EE649 pKa = 3.78FRR651 pKa = 11.84DD652 pKa = 3.37TAVTVVAGRR661 pKa = 11.84PVGLPNGAHH670 pKa = 6.39NAYY673 pKa = 10.26DD674 pKa = 3.66SQARR678 pKa = 11.84RR679 pKa = 11.84ARR681 pKa = 11.84TRR683 pKa = 11.84ATRR686 pKa = 11.84EE687 pKa = 3.73LAASAARR694 pKa = 11.84ARR696 pKa = 11.84LYY698 pKa = 11.07GRR700 pKa = 11.84ATVAEE705 pKa = 4.27MPILTSAPVLRR716 pKa = 11.84AAPPRR721 pKa = 11.84PEE723 pKa = 4.19AVNPMEE729 pKa = 4.98GGGPGHH735 pKa = 7.34DD736 pKa = 4.0LARR739 pKa = 11.84HH740 pKa = 5.42SGMGRR745 pKa = 11.84HH746 pKa = 5.96TYY748 pKa = 10.23PPDD751 pKa = 3.53RR752 pKa = 11.84EE753 pKa = 4.28QRR755 pKa = 11.84GDD757 pKa = 3.43PAVPVPQHH765 pKa = 4.83QALRR769 pKa = 11.84APQMPRR775 pKa = 11.84PGPNPQGGGVIPPPPPLPPSGGGDD799 pKa = 3.33GSGPSSGGSCAQEE812 pKa = 4.32DD813 pKa = 4.8TNPAPPVAPGPAQPDD828 pKa = 4.06GPANEE833 pKa = 4.18
Molecular weight: 88.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9YXE7|Q9YXE7_9VIRU Coat protein OS=Sphaeropsis sapinea RNA virus 1 OX=73497 PE=4 SV=1
MM1 pKa = 7.45NKK3 pKa = 10.13FIDD6 pKa = 3.68RR7 pKa = 11.84AEE9 pKa = 4.01QFGPVGDD16 pKa = 4.13YY17 pKa = 10.88LLSVVEE23 pKa = 4.5RR24 pKa = 11.84FPTVHH29 pKa = 6.84SYY31 pKa = 11.91GEE33 pKa = 4.07LSFVDD38 pKa = 3.17ALTRR42 pKa = 11.84LRR44 pKa = 11.84AMSNNLHH51 pKa = 6.81ANDD54 pKa = 4.67ALLPAAVSLLCLPFPLQVEE73 pKa = 4.51MSRR76 pKa = 11.84ADD78 pKa = 3.55VYY80 pKa = 11.78ALLVKK85 pKa = 9.59GTSLPFNFVRR95 pKa = 11.84STAHH99 pKa = 6.63LHH101 pKa = 6.01PLPDD105 pKa = 3.81DD106 pKa = 3.65VGRR109 pKa = 11.84PGSTRR114 pKa = 11.84YY115 pKa = 9.69SGRR118 pKa = 11.84FFARR122 pKa = 11.84LLNDD126 pKa = 2.99HH127 pKa = 6.29SFRR130 pKa = 11.84RR131 pKa = 11.84SCFPTKK137 pKa = 10.58KK138 pKa = 9.53MLPAEE143 pKa = 4.35VKK145 pKa = 10.36PNLALAPLIRR155 pKa = 11.84SFSTCAGPTLAGHH168 pKa = 6.73FLRR171 pKa = 11.84HH172 pKa = 5.67LAGHH176 pKa = 4.86VTEE179 pKa = 5.02DD180 pKa = 2.97VAMAAMVYY188 pKa = 10.54AHH190 pKa = 7.0GLVACYY196 pKa = 9.34GQKK199 pKa = 10.76GYY201 pKa = 10.72DD202 pKa = 3.01IAAWLVLRR210 pKa = 11.84PAAAKK215 pKa = 10.45GISTAIKK222 pKa = 9.99ALGANAHH229 pKa = 6.67DD230 pKa = 5.18FGAVLCEE237 pKa = 3.71AQTLLGRR244 pKa = 11.84AVSTIDD250 pKa = 3.43LAHH253 pKa = 6.03EE254 pKa = 4.08AEE256 pKa = 4.52YY257 pKa = 10.9RR258 pKa = 11.84CDD260 pKa = 3.74PDD262 pKa = 5.2LVAKK266 pKa = 9.51QVIEE270 pKa = 4.23PGEE273 pKa = 4.04EE274 pKa = 3.88LRR276 pKa = 11.84SHH278 pKa = 6.03IRR280 pKa = 11.84AVLAMEE286 pKa = 4.5LAGRR290 pKa = 11.84DD291 pKa = 3.62LSLPDD296 pKa = 4.28LDD298 pKa = 4.68SWWSSRR304 pKa = 11.84WLWCVNGSQNDD315 pKa = 3.4ASSRR319 pKa = 11.84LLGIDD324 pKa = 3.01TARR327 pKa = 11.84FRR329 pKa = 11.84EE330 pKa = 4.13FHH332 pKa = 5.07TRR334 pKa = 11.84EE335 pKa = 3.9YY336 pKa = 11.11RR337 pKa = 11.84RR338 pKa = 11.84MASEE342 pKa = 4.4ALTHH346 pKa = 6.23EE347 pKa = 5.76PITSWDD353 pKa = 3.49GYY355 pKa = 7.88TNISASPKK363 pKa = 9.75LEE365 pKa = 4.18HH366 pKa = 6.54GKK368 pKa = 7.98TRR370 pKa = 11.84AIFACDD376 pKa = 3.1TRR378 pKa = 11.84SYY380 pKa = 11.07FAFEE384 pKa = 4.1WLLGATQKK392 pKa = 10.68AWRR395 pKa = 11.84NHH397 pKa = 5.39RR398 pKa = 11.84VLLDD402 pKa = 3.81PGGGGHH408 pKa = 7.34LGISRR413 pKa = 11.84RR414 pKa = 11.84VRR416 pKa = 11.84SFMKK420 pKa = 10.36HH421 pKa = 5.01GGVNLMLDD429 pKa = 3.53FDD431 pKa = 5.41DD432 pKa = 5.75FNSHH436 pKa = 6.89HH437 pKa = 6.62SLNSQRR443 pKa = 11.84MLFGEE448 pKa = 4.29LCDD451 pKa = 3.95RR452 pKa = 11.84ANAPGWYY459 pKa = 9.23RR460 pKa = 11.84KK461 pKa = 9.7VLSDD465 pKa = 3.05SWGKK469 pKa = 7.92MHH471 pKa = 7.04VNIAGHH477 pKa = 4.98MRR479 pKa = 11.84PWLGTLPSGHH489 pKa = 7.16RR490 pKa = 11.84GTTIVNSVLNAAYY503 pKa = 9.67IRR505 pKa = 11.84MALGGPAFDD514 pKa = 5.23KK515 pKa = 10.34LTSLHH520 pKa = 6.47TGDD523 pKa = 4.85DD524 pKa = 3.79VYY526 pKa = 11.3IRR528 pKa = 11.84ADD530 pKa = 3.4TLTSCEE536 pKa = 4.21WILDD540 pKa = 3.61RR541 pKa = 11.84VRR543 pKa = 11.84SVGCRR548 pKa = 11.84INPAKK553 pKa = 10.39QSVGFGTGEE562 pKa = 4.26FLRR565 pKa = 11.84MAITQRR571 pKa = 11.84EE572 pKa = 4.18TRR574 pKa = 11.84GYY576 pKa = 9.74LARR579 pKa = 11.84SVASFVSGNWTNQNPLDD596 pKa = 4.22PADD599 pKa = 4.17ALRR602 pKa = 11.84TAIVSTRR609 pKa = 11.84ALINRR614 pKa = 11.84SGRR617 pKa = 11.84QDD619 pKa = 3.3YY620 pKa = 11.13ARR622 pKa = 11.84LVGPALRR629 pKa = 11.84LSHH632 pKa = 7.16PLGTRR637 pKa = 11.84NIIRR641 pKa = 11.84LLSGDD646 pKa = 3.56VALDD650 pKa = 3.9DD651 pKa = 4.42GPVYY655 pKa = 10.26NTDD658 pKa = 2.56GRR660 pKa = 11.84IVHH663 pKa = 6.23FRR665 pKa = 11.84VEE667 pKa = 4.21GQIEE671 pKa = 4.11DD672 pKa = 3.92ALPVSEE678 pKa = 4.82RR679 pKa = 11.84WPHH682 pKa = 5.68NATDD686 pKa = 4.35AYY688 pKa = 10.53LLSHH692 pKa = 6.09VSEE695 pKa = 4.59VEE697 pKa = 3.97RR698 pKa = 11.84AALAATGTDD707 pKa = 3.18AAALMLASSFSKK719 pKa = 10.24GHH721 pKa = 6.2SSLGSARR728 pKa = 11.84RR729 pKa = 11.84QVVRR733 pKa = 11.84VAKK736 pKa = 10.54RR737 pKa = 11.84EE738 pKa = 3.91TTRR741 pKa = 11.84AHH743 pKa = 6.41GFADD747 pKa = 4.47ASDD750 pKa = 3.9LLHH753 pKa = 7.48RR754 pKa = 11.84DD755 pKa = 3.17RR756 pKa = 11.84DD757 pKa = 3.85KK758 pKa = 11.87VGVLKK763 pKa = 10.65AYY765 pKa = 9.85PLVNLVKK772 pKa = 10.53NRR774 pKa = 11.84LRR776 pKa = 11.84KK777 pKa = 8.33EE778 pKa = 4.36TISEE782 pKa = 4.03LLILAGHH789 pKa = 6.23TPGADD794 pKa = 3.64PYY796 pKa = 9.11RR797 pKa = 11.84QAFGEE802 pKa = 4.21HH803 pKa = 7.0PEE805 pKa = 4.15SKK807 pKa = 10.7NIIGSLSHH815 pKa = 6.91SDD817 pKa = 3.09ASALGKK823 pKa = 9.07ATRR826 pKa = 11.84SGNIYY831 pKa = 8.43TLTPVFVV838 pKa = 3.75
MM1 pKa = 7.45NKK3 pKa = 10.13FIDD6 pKa = 3.68RR7 pKa = 11.84AEE9 pKa = 4.01QFGPVGDD16 pKa = 4.13YY17 pKa = 10.88LLSVVEE23 pKa = 4.5RR24 pKa = 11.84FPTVHH29 pKa = 6.84SYY31 pKa = 11.91GEE33 pKa = 4.07LSFVDD38 pKa = 3.17ALTRR42 pKa = 11.84LRR44 pKa = 11.84AMSNNLHH51 pKa = 6.81ANDD54 pKa = 4.67ALLPAAVSLLCLPFPLQVEE73 pKa = 4.51MSRR76 pKa = 11.84ADD78 pKa = 3.55VYY80 pKa = 11.78ALLVKK85 pKa = 9.59GTSLPFNFVRR95 pKa = 11.84STAHH99 pKa = 6.63LHH101 pKa = 6.01PLPDD105 pKa = 3.81DD106 pKa = 3.65VGRR109 pKa = 11.84PGSTRR114 pKa = 11.84YY115 pKa = 9.69SGRR118 pKa = 11.84FFARR122 pKa = 11.84LLNDD126 pKa = 2.99HH127 pKa = 6.29SFRR130 pKa = 11.84RR131 pKa = 11.84SCFPTKK137 pKa = 10.58KK138 pKa = 9.53MLPAEE143 pKa = 4.35VKK145 pKa = 10.36PNLALAPLIRR155 pKa = 11.84SFSTCAGPTLAGHH168 pKa = 6.73FLRR171 pKa = 11.84HH172 pKa = 5.67LAGHH176 pKa = 4.86VTEE179 pKa = 5.02DD180 pKa = 2.97VAMAAMVYY188 pKa = 10.54AHH190 pKa = 7.0GLVACYY196 pKa = 9.34GQKK199 pKa = 10.76GYY201 pKa = 10.72DD202 pKa = 3.01IAAWLVLRR210 pKa = 11.84PAAAKK215 pKa = 10.45GISTAIKK222 pKa = 9.99ALGANAHH229 pKa = 6.67DD230 pKa = 5.18FGAVLCEE237 pKa = 3.71AQTLLGRR244 pKa = 11.84AVSTIDD250 pKa = 3.43LAHH253 pKa = 6.03EE254 pKa = 4.08AEE256 pKa = 4.52YY257 pKa = 10.9RR258 pKa = 11.84CDD260 pKa = 3.74PDD262 pKa = 5.2LVAKK266 pKa = 9.51QVIEE270 pKa = 4.23PGEE273 pKa = 4.04EE274 pKa = 3.88LRR276 pKa = 11.84SHH278 pKa = 6.03IRR280 pKa = 11.84AVLAMEE286 pKa = 4.5LAGRR290 pKa = 11.84DD291 pKa = 3.62LSLPDD296 pKa = 4.28LDD298 pKa = 4.68SWWSSRR304 pKa = 11.84WLWCVNGSQNDD315 pKa = 3.4ASSRR319 pKa = 11.84LLGIDD324 pKa = 3.01TARR327 pKa = 11.84FRR329 pKa = 11.84EE330 pKa = 4.13FHH332 pKa = 5.07TRR334 pKa = 11.84EE335 pKa = 3.9YY336 pKa = 11.11RR337 pKa = 11.84RR338 pKa = 11.84MASEE342 pKa = 4.4ALTHH346 pKa = 6.23EE347 pKa = 5.76PITSWDD353 pKa = 3.49GYY355 pKa = 7.88TNISASPKK363 pKa = 9.75LEE365 pKa = 4.18HH366 pKa = 6.54GKK368 pKa = 7.98TRR370 pKa = 11.84AIFACDD376 pKa = 3.1TRR378 pKa = 11.84SYY380 pKa = 11.07FAFEE384 pKa = 4.1WLLGATQKK392 pKa = 10.68AWRR395 pKa = 11.84NHH397 pKa = 5.39RR398 pKa = 11.84VLLDD402 pKa = 3.81PGGGGHH408 pKa = 7.34LGISRR413 pKa = 11.84RR414 pKa = 11.84VRR416 pKa = 11.84SFMKK420 pKa = 10.36HH421 pKa = 5.01GGVNLMLDD429 pKa = 3.53FDD431 pKa = 5.41DD432 pKa = 5.75FNSHH436 pKa = 6.89HH437 pKa = 6.62SLNSQRR443 pKa = 11.84MLFGEE448 pKa = 4.29LCDD451 pKa = 3.95RR452 pKa = 11.84ANAPGWYY459 pKa = 9.23RR460 pKa = 11.84KK461 pKa = 9.7VLSDD465 pKa = 3.05SWGKK469 pKa = 7.92MHH471 pKa = 7.04VNIAGHH477 pKa = 4.98MRR479 pKa = 11.84PWLGTLPSGHH489 pKa = 7.16RR490 pKa = 11.84GTTIVNSVLNAAYY503 pKa = 9.67IRR505 pKa = 11.84MALGGPAFDD514 pKa = 5.23KK515 pKa = 10.34LTSLHH520 pKa = 6.47TGDD523 pKa = 4.85DD524 pKa = 3.79VYY526 pKa = 11.3IRR528 pKa = 11.84ADD530 pKa = 3.4TLTSCEE536 pKa = 4.21WILDD540 pKa = 3.61RR541 pKa = 11.84VRR543 pKa = 11.84SVGCRR548 pKa = 11.84INPAKK553 pKa = 10.39QSVGFGTGEE562 pKa = 4.26FLRR565 pKa = 11.84MAITQRR571 pKa = 11.84EE572 pKa = 4.18TRR574 pKa = 11.84GYY576 pKa = 9.74LARR579 pKa = 11.84SVASFVSGNWTNQNPLDD596 pKa = 4.22PADD599 pKa = 4.17ALRR602 pKa = 11.84TAIVSTRR609 pKa = 11.84ALINRR614 pKa = 11.84SGRR617 pKa = 11.84QDD619 pKa = 3.3YY620 pKa = 11.13ARR622 pKa = 11.84LVGPALRR629 pKa = 11.84LSHH632 pKa = 7.16PLGTRR637 pKa = 11.84NIIRR641 pKa = 11.84LLSGDD646 pKa = 3.56VALDD650 pKa = 3.9DD651 pKa = 4.42GPVYY655 pKa = 10.26NTDD658 pKa = 2.56GRR660 pKa = 11.84IVHH663 pKa = 6.23FRR665 pKa = 11.84VEE667 pKa = 4.21GQIEE671 pKa = 4.11DD672 pKa = 3.92ALPVSEE678 pKa = 4.82RR679 pKa = 11.84WPHH682 pKa = 5.68NATDD686 pKa = 4.35AYY688 pKa = 10.53LLSHH692 pKa = 6.09VSEE695 pKa = 4.59VEE697 pKa = 3.97RR698 pKa = 11.84AALAATGTDD707 pKa = 3.18AAALMLASSFSKK719 pKa = 10.24GHH721 pKa = 6.2SSLGSARR728 pKa = 11.84RR729 pKa = 11.84QVVRR733 pKa = 11.84VAKK736 pKa = 10.54RR737 pKa = 11.84EE738 pKa = 3.91TTRR741 pKa = 11.84AHH743 pKa = 6.41GFADD747 pKa = 4.47ASDD750 pKa = 3.9LLHH753 pKa = 7.48RR754 pKa = 11.84DD755 pKa = 3.17RR756 pKa = 11.84DD757 pKa = 3.85KK758 pKa = 11.87VGVLKK763 pKa = 10.65AYY765 pKa = 9.85PLVNLVKK772 pKa = 10.53NRR774 pKa = 11.84LRR776 pKa = 11.84KK777 pKa = 8.33EE778 pKa = 4.36TISEE782 pKa = 4.03LLILAGHH789 pKa = 6.23TPGADD794 pKa = 3.64PYY796 pKa = 9.11RR797 pKa = 11.84QAFGEE802 pKa = 4.21HH803 pKa = 7.0PEE805 pKa = 4.15SKK807 pKa = 10.7NIIGSLSHH815 pKa = 6.91SDD817 pKa = 3.09ASALGKK823 pKa = 9.07ATRR826 pKa = 11.84SGNIYY831 pKa = 8.43TLTPVFVV838 pKa = 3.75
Molecular weight: 92.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1671 |
833 |
838 |
835.5 |
90.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.55 ± 0.432 | 1.197 ± 0.088 |
5.386 ± 0.259 | 4.249 ± 0.235 |
3.83 ± 0.009 | 8.917 ± 0.878 |
3.291 ± 0.579 | 3.71 ± 0.008 |
1.915 ± 0.808 | 10.293 ± 0.88 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.855 ± 0.041 | 3.591 ± 0.082 |
7.121 ± 1.957 | 2.214 ± 0.321 |
8.259 ± 0.162 | 6.822 ± 0.797 |
5.685 ± 0.239 | 6.463 ± 0.015 |
1.257 ± 0.313 | 2.394 ± 0.085 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
