
Aerococcus viridans ATCC 11563 = CCUG 4311
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Aerococcaceae; Aerococcus; Aerococcus viridans
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
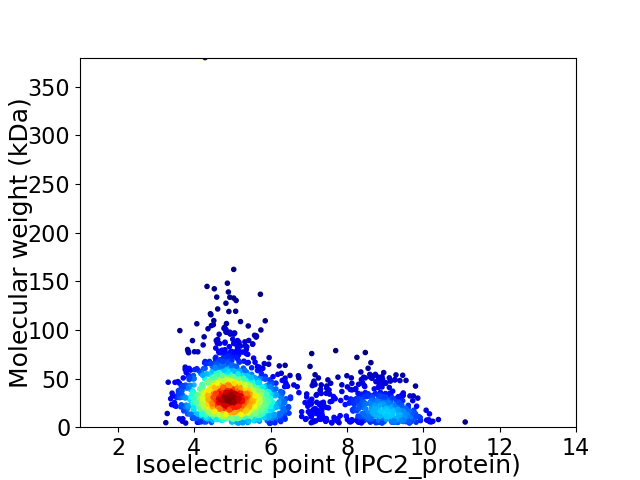
Virtual 2D-PAGE plot for 1929 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4YEF0|D4YEF0_9LACT Uncharacterized protein OS=Aerococcus viridans ATCC 11563 = CCUG 4311 OX=655812 GN=HMPREF0061_0239 PE=4 SV=1
MM1 pKa = 7.21NKK3 pKa = 10.08KK4 pKa = 10.48LGILSLTALSLVLAACGNGGTTADD28 pKa = 3.77NEE30 pKa = 4.13EE31 pKa = 3.92AAKK34 pKa = 10.31SIKK37 pKa = 9.31VTVNGEE43 pKa = 4.03LASLDD48 pKa = 4.42SISSTDD54 pKa = 3.41SPSQNTIANVMEE66 pKa = 4.57GLYY69 pKa = 10.35RR70 pKa = 11.84PADD73 pKa = 3.6NGSNEE78 pKa = 4.2LGVAAEE84 pKa = 4.41EE85 pKa = 4.42PTVSEE90 pKa = 5.32DD91 pKa = 2.92GLTYY95 pKa = 10.63TFTIRR100 pKa = 11.84DD101 pKa = 3.66DD102 pKa = 4.67ADD104 pKa = 3.23WSNGDD109 pKa = 3.88PVTAQDD115 pKa = 5.14FVYY118 pKa = 9.49TYY120 pKa = 10.47QKK122 pKa = 9.56TVTPEE127 pKa = 4.2TIGGNANKK135 pKa = 10.14FYY137 pKa = 10.21MIEE140 pKa = 3.91NAEE143 pKa = 4.35AIANGEE149 pKa = 4.23AEE151 pKa = 4.56PDD153 pKa = 3.65TLGVKK158 pKa = 10.5AIDD161 pKa = 4.1DD162 pKa = 3.73KK163 pKa = 11.47TLEE166 pKa = 4.17FTLASPTTYY175 pKa = 8.96FTDD178 pKa = 4.24LLATPYY184 pKa = 10.14YY185 pKa = 10.81LPVNHH190 pKa = 6.3NVATEE195 pKa = 4.17LGSDD199 pKa = 3.53YY200 pKa = 10.31GTSAEE205 pKa = 3.96NAVYY209 pKa = 10.46NGPFEE214 pKa = 4.3VSDD217 pKa = 3.65WNSTEE222 pKa = 4.51IEE224 pKa = 3.92WTLTKK229 pKa = 10.52NDD231 pKa = 4.44EE232 pKa = 4.08YY233 pKa = 10.95WDD235 pKa = 4.07ADD237 pKa = 4.07NVDD240 pKa = 4.6LDD242 pKa = 3.98QIDD245 pKa = 3.37WVMSKK250 pKa = 10.74EE251 pKa = 3.77NSTNVNLFEE260 pKa = 5.84GGEE263 pKa = 3.98LQYY266 pKa = 11.19TEE268 pKa = 3.42ITAPYY273 pKa = 8.27VRR275 pKa = 11.84EE276 pKa = 4.02YY277 pKa = 11.12QDD279 pKa = 2.97TDD281 pKa = 3.54VLHH284 pKa = 6.98IEE286 pKa = 4.27PKK288 pKa = 10.8GMTGYY293 pKa = 9.46MVFNTQSSPTNNVHH307 pKa = 6.14FRR309 pKa = 11.84RR310 pKa = 11.84AISQAYY316 pKa = 9.53DD317 pKa = 2.93KK318 pKa = 10.83EE319 pKa = 4.52AFVDD323 pKa = 4.23AVLDD327 pKa = 4.72DD328 pKa = 4.58GSQAAGGWIPSDD340 pKa = 3.58FGSDD344 pKa = 3.61PEE346 pKa = 4.22SGIDD350 pKa = 3.61FRR352 pKa = 11.84EE353 pKa = 4.09EE354 pKa = 3.68NGDD357 pKa = 4.21LMTFDD362 pKa = 5.35LEE364 pKa = 4.3AAQEE368 pKa = 4.03EE369 pKa = 4.7WALALEE375 pKa = 4.33EE376 pKa = 5.16LGTDD380 pKa = 3.35TVEE383 pKa = 4.91IEE385 pKa = 4.78LLTSDD390 pKa = 4.06TDD392 pKa = 3.56TSKK395 pKa = 10.33ATSEE399 pKa = 3.96YY400 pKa = 10.83LQAQLQTNLPGLTVTVRR417 pKa = 11.84NVPLKK422 pKa = 10.49SRR424 pKa = 11.84QAITGTGEE432 pKa = 3.44FDD434 pKa = 3.47IVYY437 pKa = 8.4GTYY440 pKa = 10.05NPSYY444 pKa = 10.96ADD446 pKa = 3.23PTAFLDD452 pKa = 4.25FFITGSPLSSGDD464 pKa = 3.8FSDD467 pKa = 3.35ATYY470 pKa = 11.07DD471 pKa = 3.52EE472 pKa = 5.47LMTEE476 pKa = 4.62GKK478 pKa = 7.96ITNALDD484 pKa = 3.61PAARR488 pKa = 11.84WDD490 pKa = 3.68NFLAAEE496 pKa = 4.54KK497 pKa = 10.97YY498 pKa = 10.32LVEE501 pKa = 4.65DD502 pKa = 4.11AAAVSPIYY510 pKa = 10.39QGAYY514 pKa = 10.47ANLIDD519 pKa = 4.39PALEE523 pKa = 3.97NVINQPNGVAQYY535 pKa = 10.79YY536 pKa = 9.5RR537 pKa = 11.84AATYY541 pKa = 10.9NVTEE545 pKa = 4.17
MM1 pKa = 7.21NKK3 pKa = 10.08KK4 pKa = 10.48LGILSLTALSLVLAACGNGGTTADD28 pKa = 3.77NEE30 pKa = 4.13EE31 pKa = 3.92AAKK34 pKa = 10.31SIKK37 pKa = 9.31VTVNGEE43 pKa = 4.03LASLDD48 pKa = 4.42SISSTDD54 pKa = 3.41SPSQNTIANVMEE66 pKa = 4.57GLYY69 pKa = 10.35RR70 pKa = 11.84PADD73 pKa = 3.6NGSNEE78 pKa = 4.2LGVAAEE84 pKa = 4.41EE85 pKa = 4.42PTVSEE90 pKa = 5.32DD91 pKa = 2.92GLTYY95 pKa = 10.63TFTIRR100 pKa = 11.84DD101 pKa = 3.66DD102 pKa = 4.67ADD104 pKa = 3.23WSNGDD109 pKa = 3.88PVTAQDD115 pKa = 5.14FVYY118 pKa = 9.49TYY120 pKa = 10.47QKK122 pKa = 9.56TVTPEE127 pKa = 4.2TIGGNANKK135 pKa = 10.14FYY137 pKa = 10.21MIEE140 pKa = 3.91NAEE143 pKa = 4.35AIANGEE149 pKa = 4.23AEE151 pKa = 4.56PDD153 pKa = 3.65TLGVKK158 pKa = 10.5AIDD161 pKa = 4.1DD162 pKa = 3.73KK163 pKa = 11.47TLEE166 pKa = 4.17FTLASPTTYY175 pKa = 8.96FTDD178 pKa = 4.24LLATPYY184 pKa = 10.14YY185 pKa = 10.81LPVNHH190 pKa = 6.3NVATEE195 pKa = 4.17LGSDD199 pKa = 3.53YY200 pKa = 10.31GTSAEE205 pKa = 3.96NAVYY209 pKa = 10.46NGPFEE214 pKa = 4.3VSDD217 pKa = 3.65WNSTEE222 pKa = 4.51IEE224 pKa = 3.92WTLTKK229 pKa = 10.52NDD231 pKa = 4.44EE232 pKa = 4.08YY233 pKa = 10.95WDD235 pKa = 4.07ADD237 pKa = 4.07NVDD240 pKa = 4.6LDD242 pKa = 3.98QIDD245 pKa = 3.37WVMSKK250 pKa = 10.74EE251 pKa = 3.77NSTNVNLFEE260 pKa = 5.84GGEE263 pKa = 3.98LQYY266 pKa = 11.19TEE268 pKa = 3.42ITAPYY273 pKa = 8.27VRR275 pKa = 11.84EE276 pKa = 4.02YY277 pKa = 11.12QDD279 pKa = 2.97TDD281 pKa = 3.54VLHH284 pKa = 6.98IEE286 pKa = 4.27PKK288 pKa = 10.8GMTGYY293 pKa = 9.46MVFNTQSSPTNNVHH307 pKa = 6.14FRR309 pKa = 11.84RR310 pKa = 11.84AISQAYY316 pKa = 9.53DD317 pKa = 2.93KK318 pKa = 10.83EE319 pKa = 4.52AFVDD323 pKa = 4.23AVLDD327 pKa = 4.72DD328 pKa = 4.58GSQAAGGWIPSDD340 pKa = 3.58FGSDD344 pKa = 3.61PEE346 pKa = 4.22SGIDD350 pKa = 3.61FRR352 pKa = 11.84EE353 pKa = 4.09EE354 pKa = 3.68NGDD357 pKa = 4.21LMTFDD362 pKa = 5.35LEE364 pKa = 4.3AAQEE368 pKa = 4.03EE369 pKa = 4.7WALALEE375 pKa = 4.33EE376 pKa = 5.16LGTDD380 pKa = 3.35TVEE383 pKa = 4.91IEE385 pKa = 4.78LLTSDD390 pKa = 4.06TDD392 pKa = 3.56TSKK395 pKa = 10.33ATSEE399 pKa = 3.96YY400 pKa = 10.83LQAQLQTNLPGLTVTVRR417 pKa = 11.84NVPLKK422 pKa = 10.49SRR424 pKa = 11.84QAITGTGEE432 pKa = 3.44FDD434 pKa = 3.47IVYY437 pKa = 8.4GTYY440 pKa = 10.05NPSYY444 pKa = 10.96ADD446 pKa = 3.23PTAFLDD452 pKa = 4.25FFITGSPLSSGDD464 pKa = 3.8FSDD467 pKa = 3.35ATYY470 pKa = 11.07DD471 pKa = 3.52EE472 pKa = 5.47LMTEE476 pKa = 4.62GKK478 pKa = 7.96ITNALDD484 pKa = 3.61PAARR488 pKa = 11.84WDD490 pKa = 3.68NFLAAEE496 pKa = 4.54KK497 pKa = 10.97YY498 pKa = 10.32LVEE501 pKa = 4.65DD502 pKa = 4.11AAAVSPIYY510 pKa = 10.39QGAYY514 pKa = 10.47ANLIDD519 pKa = 4.39PALEE523 pKa = 3.97NVINQPNGVAQYY535 pKa = 10.79YY536 pKa = 9.5RR537 pKa = 11.84AATYY541 pKa = 10.9NVTEE545 pKa = 4.17
Molecular weight: 59.37 kDa
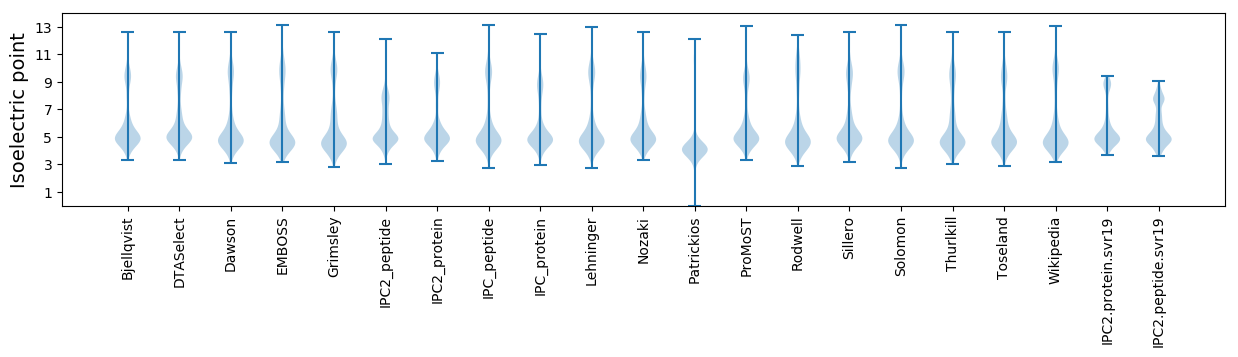
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4YHD1|D4YHD1_9LACT Thioredoxin-like_fold domain-containing protein OS=Aerococcus viridans ATCC 11563 = CCUG 4311 OX=655812 GN=HMPREF0061_1270 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 9.2KK9 pKa = 7.96RR10 pKa = 11.84HH11 pKa = 4.8RR12 pKa = 11.84QKK14 pKa = 10.16VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 10.16NGRR28 pKa = 11.84HH29 pKa = 4.13VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.53GRR39 pKa = 11.84KK40 pKa = 9.06VISAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 9.2KK9 pKa = 7.96RR10 pKa = 11.84HH11 pKa = 4.8RR12 pKa = 11.84QKK14 pKa = 10.16VHH16 pKa = 5.49GFRR19 pKa = 11.84KK20 pKa = 10.04RR21 pKa = 11.84MSTKK25 pKa = 10.16NGRR28 pKa = 11.84HH29 pKa = 4.13VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.53GRR39 pKa = 11.84KK40 pKa = 9.06VISAA44 pKa = 4.05
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
571387 |
37 |
3427 |
296.2 |
33.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.115 ± 0.067 | 0.436 ± 0.014 |
6.407 ± 0.042 | 7.141 ± 0.064 |
4.298 ± 0.043 | 6.733 ± 0.057 |
1.896 ± 0.028 | 7.594 ± 0.054 |
5.821 ± 0.052 | 9.096 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.725 ± 0.029 | 4.848 ± 0.041 |
3.394 ± 0.034 | 4.335 ± 0.052 |
3.947 ± 0.042 | 5.76 ± 0.056 |
5.746 ± 0.038 | 7.102 ± 0.045 |
0.847 ± 0.018 | 3.758 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
