
Nilaparvata lugens reovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Fijivirus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
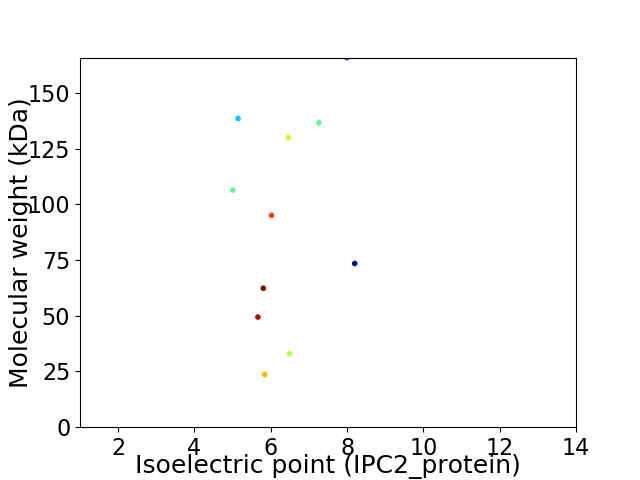
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q83863|Q83863_9REOV 95.1KD putative nonstructural protein OS=Nilaparvata lugens reovirus OX=33724 PE=4 SV=1
MM1 pKa = 7.61AFKK4 pKa = 10.51FDD6 pKa = 3.95ASNLNKK12 pKa = 9.24TRR14 pKa = 11.84QALATMSNPSFQPKK28 pKa = 9.85GFPSNSTQFNGSNGRR43 pKa = 11.84KK44 pKa = 9.62GKK46 pKa = 10.53FPDD49 pKa = 3.5AKK51 pKa = 9.62KK52 pKa = 7.26TTLSSPPLFNEE63 pKa = 3.8SQKK66 pKa = 10.77PKK68 pKa = 9.31TVSSMITSPIIGSDD82 pKa = 3.72RR83 pKa = 11.84IFTSSEE89 pKa = 3.91FIHH92 pKa = 5.8PQKK95 pKa = 10.74FVMTEE100 pKa = 3.78YY101 pKa = 9.69VQKK104 pKa = 10.86SIAKK108 pKa = 9.27FEE110 pKa = 4.27THH112 pKa = 6.7GSIDD116 pKa = 3.83PFAKK120 pKa = 10.52KK121 pKa = 9.88PDD123 pKa = 3.82FDD125 pKa = 4.62DD126 pKa = 4.19LCLILYY132 pKa = 7.74TPAASKK138 pKa = 11.08HH139 pKa = 5.08EE140 pKa = 4.18LSLANAQLVCSDD152 pKa = 3.26IDD154 pKa = 3.81SLNLHH159 pKa = 6.14SMHH162 pKa = 6.88VIDD165 pKa = 4.04GTKK168 pKa = 9.89LYY170 pKa = 11.48SMAANVQYY178 pKa = 11.25LNAGVIYY185 pKa = 10.07LPKK188 pKa = 9.96SHH190 pKa = 7.1WDD192 pKa = 3.02AFTSKK197 pKa = 7.69THH199 pKa = 5.79AHH201 pKa = 5.92VKK203 pKa = 9.76MEE205 pKa = 4.27MNFACSPVHH214 pKa = 6.54LNKK217 pKa = 10.14LRR219 pKa = 11.84LAHH222 pKa = 6.17WMSPFLLTKK231 pKa = 10.22MSVSFQLQVEE241 pKa = 4.64IYY243 pKa = 10.45DD244 pKa = 3.65HH245 pKa = 7.2FEE247 pKa = 3.86QFVKK251 pKa = 9.62NTHH254 pKa = 6.47ANVDD258 pKa = 3.17SPMRR262 pKa = 11.84VFGSKK267 pKa = 9.78NGKK270 pKa = 8.78EE271 pKa = 4.29CGSLEE276 pKa = 4.14PEE278 pKa = 4.34EE279 pKa = 5.75DD280 pKa = 3.68LDD282 pKa = 5.06SPLDD286 pKa = 3.71FSKK289 pKa = 11.59NNVCLIDD296 pKa = 3.82VNDD299 pKa = 4.7DD300 pKa = 3.26FTSAEE305 pKa = 3.8RR306 pKa = 11.84KK307 pKa = 7.41MHH309 pKa = 5.8VNLNVFRR316 pKa = 11.84NAITFVKK323 pKa = 10.59KK324 pKa = 10.51EE325 pKa = 4.07FGRR328 pKa = 11.84ANGGNSLVQKK338 pKa = 9.72MRR340 pKa = 11.84ASKK343 pKa = 10.9VFLLYY348 pKa = 10.44FDD350 pKa = 5.62LNNTSEE356 pKa = 4.11TRR358 pKa = 11.84NRR360 pKa = 11.84KK361 pKa = 8.49QRR363 pKa = 11.84DD364 pKa = 3.42PSVVNTDD371 pKa = 3.3TLKK374 pKa = 11.19AFDD377 pKa = 4.02GFNTVFVIVDD387 pKa = 3.95SEE389 pKa = 4.33GMIASSVSMSSVEE402 pKa = 4.08AMLMSLKK409 pKa = 10.22QVSLVDD415 pKa = 3.13YY416 pKa = 10.13HH417 pKa = 7.73LNYY420 pKa = 10.65CSDD423 pKa = 3.09VLMNEE428 pKa = 3.77GHH430 pKa = 6.55VYY432 pKa = 11.03SLMLSCICKK441 pKa = 8.67TLSFGGRR448 pKa = 11.84ILISSWIHH456 pKa = 5.89ALFEE460 pKa = 4.6GEE462 pKa = 4.8KK463 pKa = 9.21KK464 pKa = 8.67TATEE468 pKa = 3.97TFEE471 pKa = 4.07EE472 pKa = 4.4LKK474 pKa = 10.68KK475 pKa = 11.23LKK477 pKa = 10.45DD478 pKa = 3.83VEE480 pKa = 4.51DD481 pKa = 4.2KK482 pKa = 11.37KK483 pKa = 10.76PDD485 pKa = 3.05AMTARR490 pKa = 11.84KK491 pKa = 7.37PTTDD495 pKa = 3.13EE496 pKa = 4.56NEE498 pKa = 4.58DD499 pKa = 3.4EE500 pKa = 4.45NEE502 pKa = 4.0AAKK505 pKa = 9.26TAFDD509 pKa = 3.3KK510 pKa = 11.43HH511 pKa = 6.52LDD513 pKa = 3.55KK514 pKa = 11.47VEE516 pKa = 4.9KK517 pKa = 10.22ILSEE521 pKa = 4.29DD522 pKa = 4.24SEE524 pKa = 4.66NQDD527 pKa = 2.84GTQTDD532 pKa = 3.7TEE534 pKa = 4.18EE535 pKa = 5.24RR536 pKa = 11.84LTQTTAAPNPIEE548 pKa = 4.31TTFPAAQTQPDD559 pKa = 3.71QTQKK563 pKa = 9.77TEE565 pKa = 4.08STDD568 pKa = 3.57QNDD571 pKa = 3.89SNVQDD576 pKa = 3.72SQEE579 pKa = 4.06NQSKK583 pKa = 10.83DD584 pKa = 3.36NQTRR588 pKa = 11.84SHH590 pKa = 7.5PLPDD594 pKa = 3.52QDD596 pKa = 5.66SVDD599 pKa = 4.34DD600 pKa = 3.87STKK603 pKa = 10.77EE604 pKa = 3.79SDD606 pKa = 3.56ASSSATFDD614 pKa = 3.17SATEE618 pKa = 4.08FKK620 pKa = 10.32EE621 pKa = 3.84AANASVLVDD630 pKa = 3.92LQDD633 pKa = 3.32IFSNASSQTAQSSTTSSTKK652 pKa = 9.51STSDD656 pKa = 2.78QATFKK661 pKa = 10.32NTVSKK666 pKa = 9.32QTDD669 pKa = 3.24SRR671 pKa = 11.84SAFPDD676 pKa = 3.12IQEE679 pKa = 4.41KK680 pKa = 9.91QLPAPPNSLDD690 pKa = 3.89DD691 pKa = 5.16GISASMGLPPPKK703 pKa = 10.06IMTVDD708 pKa = 3.46EE709 pKa = 4.36VEE711 pKa = 4.08AALFGTKK718 pKa = 9.99EE719 pKa = 4.13NGSVASPVKK728 pKa = 10.31NVPIHH733 pKa = 6.27EE734 pKa = 4.37KK735 pKa = 10.68ASPKK739 pKa = 9.51NANGRR744 pKa = 11.84TLPPVPEE751 pKa = 4.18EE752 pKa = 4.06VPIGNVGGPRR762 pKa = 11.84QLKK765 pKa = 10.47VSFDD769 pKa = 3.94DD770 pKa = 6.41LMDD773 pKa = 4.44FSQNASAVQNSCPSNNIPPSPSEE796 pKa = 3.86SSEE799 pKa = 4.01TTKK802 pKa = 10.52QQSSSCGSSNNPSSSTGYY820 pKa = 10.79VGDD823 pKa = 3.8NTKK826 pKa = 9.6TEE828 pKa = 4.33PLPTTPSPSDD838 pKa = 3.21AASTKK843 pKa = 10.75SVDD846 pKa = 3.41NDD848 pKa = 3.7ALSTPPNGNVDD859 pKa = 3.45DD860 pKa = 4.96HH861 pKa = 5.72QAKK864 pKa = 10.41SNPSSVFDD872 pKa = 4.08FTASCQSSKK881 pKa = 11.02SDD883 pKa = 4.13SGLCTPTASTTSDD896 pKa = 3.23PNQALFYY903 pKa = 10.28QQFSFIKK910 pKa = 9.89EE911 pKa = 4.15YY912 pKa = 11.18NCTPEE917 pKa = 3.88NLRR920 pKa = 11.84LLLEE924 pKa = 4.23TVRR927 pKa = 11.84ARR929 pKa = 11.84LIPMHH934 pKa = 6.64LVHH937 pKa = 7.08PEE939 pKa = 3.77VVPNLDD945 pKa = 3.75SVCEE949 pKa = 3.96PRR951 pKa = 11.84NLSLAFGLSTLCYY964 pKa = 10.27DD965 pKa = 4.19PNPNTGCC972 pKa = 3.92
MM1 pKa = 7.61AFKK4 pKa = 10.51FDD6 pKa = 3.95ASNLNKK12 pKa = 9.24TRR14 pKa = 11.84QALATMSNPSFQPKK28 pKa = 9.85GFPSNSTQFNGSNGRR43 pKa = 11.84KK44 pKa = 9.62GKK46 pKa = 10.53FPDD49 pKa = 3.5AKK51 pKa = 9.62KK52 pKa = 7.26TTLSSPPLFNEE63 pKa = 3.8SQKK66 pKa = 10.77PKK68 pKa = 9.31TVSSMITSPIIGSDD82 pKa = 3.72RR83 pKa = 11.84IFTSSEE89 pKa = 3.91FIHH92 pKa = 5.8PQKK95 pKa = 10.74FVMTEE100 pKa = 3.78YY101 pKa = 9.69VQKK104 pKa = 10.86SIAKK108 pKa = 9.27FEE110 pKa = 4.27THH112 pKa = 6.7GSIDD116 pKa = 3.83PFAKK120 pKa = 10.52KK121 pKa = 9.88PDD123 pKa = 3.82FDD125 pKa = 4.62DD126 pKa = 4.19LCLILYY132 pKa = 7.74TPAASKK138 pKa = 11.08HH139 pKa = 5.08EE140 pKa = 4.18LSLANAQLVCSDD152 pKa = 3.26IDD154 pKa = 3.81SLNLHH159 pKa = 6.14SMHH162 pKa = 6.88VIDD165 pKa = 4.04GTKK168 pKa = 9.89LYY170 pKa = 11.48SMAANVQYY178 pKa = 11.25LNAGVIYY185 pKa = 10.07LPKK188 pKa = 9.96SHH190 pKa = 7.1WDD192 pKa = 3.02AFTSKK197 pKa = 7.69THH199 pKa = 5.79AHH201 pKa = 5.92VKK203 pKa = 9.76MEE205 pKa = 4.27MNFACSPVHH214 pKa = 6.54LNKK217 pKa = 10.14LRR219 pKa = 11.84LAHH222 pKa = 6.17WMSPFLLTKK231 pKa = 10.22MSVSFQLQVEE241 pKa = 4.64IYY243 pKa = 10.45DD244 pKa = 3.65HH245 pKa = 7.2FEE247 pKa = 3.86QFVKK251 pKa = 9.62NTHH254 pKa = 6.47ANVDD258 pKa = 3.17SPMRR262 pKa = 11.84VFGSKK267 pKa = 9.78NGKK270 pKa = 8.78EE271 pKa = 4.29CGSLEE276 pKa = 4.14PEE278 pKa = 4.34EE279 pKa = 5.75DD280 pKa = 3.68LDD282 pKa = 5.06SPLDD286 pKa = 3.71FSKK289 pKa = 11.59NNVCLIDD296 pKa = 3.82VNDD299 pKa = 4.7DD300 pKa = 3.26FTSAEE305 pKa = 3.8RR306 pKa = 11.84KK307 pKa = 7.41MHH309 pKa = 5.8VNLNVFRR316 pKa = 11.84NAITFVKK323 pKa = 10.59KK324 pKa = 10.51EE325 pKa = 4.07FGRR328 pKa = 11.84ANGGNSLVQKK338 pKa = 9.72MRR340 pKa = 11.84ASKK343 pKa = 10.9VFLLYY348 pKa = 10.44FDD350 pKa = 5.62LNNTSEE356 pKa = 4.11TRR358 pKa = 11.84NRR360 pKa = 11.84KK361 pKa = 8.49QRR363 pKa = 11.84DD364 pKa = 3.42PSVVNTDD371 pKa = 3.3TLKK374 pKa = 11.19AFDD377 pKa = 4.02GFNTVFVIVDD387 pKa = 3.95SEE389 pKa = 4.33GMIASSVSMSSVEE402 pKa = 4.08AMLMSLKK409 pKa = 10.22QVSLVDD415 pKa = 3.13YY416 pKa = 10.13HH417 pKa = 7.73LNYY420 pKa = 10.65CSDD423 pKa = 3.09VLMNEE428 pKa = 3.77GHH430 pKa = 6.55VYY432 pKa = 11.03SLMLSCICKK441 pKa = 8.67TLSFGGRR448 pKa = 11.84ILISSWIHH456 pKa = 5.89ALFEE460 pKa = 4.6GEE462 pKa = 4.8KK463 pKa = 9.21KK464 pKa = 8.67TATEE468 pKa = 3.97TFEE471 pKa = 4.07EE472 pKa = 4.4LKK474 pKa = 10.68KK475 pKa = 11.23LKK477 pKa = 10.45DD478 pKa = 3.83VEE480 pKa = 4.51DD481 pKa = 4.2KK482 pKa = 11.37KK483 pKa = 10.76PDD485 pKa = 3.05AMTARR490 pKa = 11.84KK491 pKa = 7.37PTTDD495 pKa = 3.13EE496 pKa = 4.56NEE498 pKa = 4.58DD499 pKa = 3.4EE500 pKa = 4.45NEE502 pKa = 4.0AAKK505 pKa = 9.26TAFDD509 pKa = 3.3KK510 pKa = 11.43HH511 pKa = 6.52LDD513 pKa = 3.55KK514 pKa = 11.47VEE516 pKa = 4.9KK517 pKa = 10.22ILSEE521 pKa = 4.29DD522 pKa = 4.24SEE524 pKa = 4.66NQDD527 pKa = 2.84GTQTDD532 pKa = 3.7TEE534 pKa = 4.18EE535 pKa = 5.24RR536 pKa = 11.84LTQTTAAPNPIEE548 pKa = 4.31TTFPAAQTQPDD559 pKa = 3.71QTQKK563 pKa = 9.77TEE565 pKa = 4.08STDD568 pKa = 3.57QNDD571 pKa = 3.89SNVQDD576 pKa = 3.72SQEE579 pKa = 4.06NQSKK583 pKa = 10.83DD584 pKa = 3.36NQTRR588 pKa = 11.84SHH590 pKa = 7.5PLPDD594 pKa = 3.52QDD596 pKa = 5.66SVDD599 pKa = 4.34DD600 pKa = 3.87STKK603 pKa = 10.77EE604 pKa = 3.79SDD606 pKa = 3.56ASSSATFDD614 pKa = 3.17SATEE618 pKa = 4.08FKK620 pKa = 10.32EE621 pKa = 3.84AANASVLVDD630 pKa = 3.92LQDD633 pKa = 3.32IFSNASSQTAQSSTTSSTKK652 pKa = 9.51STSDD656 pKa = 2.78QATFKK661 pKa = 10.32NTVSKK666 pKa = 9.32QTDD669 pKa = 3.24SRR671 pKa = 11.84SAFPDD676 pKa = 3.12IQEE679 pKa = 4.41KK680 pKa = 9.91QLPAPPNSLDD690 pKa = 3.89DD691 pKa = 5.16GISASMGLPPPKK703 pKa = 10.06IMTVDD708 pKa = 3.46EE709 pKa = 4.36VEE711 pKa = 4.08AALFGTKK718 pKa = 9.99EE719 pKa = 4.13NGSVASPVKK728 pKa = 10.31NVPIHH733 pKa = 6.27EE734 pKa = 4.37KK735 pKa = 10.68ASPKK739 pKa = 9.51NANGRR744 pKa = 11.84TLPPVPEE751 pKa = 4.18EE752 pKa = 4.06VPIGNVGGPRR762 pKa = 11.84QLKK765 pKa = 10.47VSFDD769 pKa = 3.94DD770 pKa = 6.41LMDD773 pKa = 4.44FSQNASAVQNSCPSNNIPPSPSEE796 pKa = 3.86SSEE799 pKa = 4.01TTKK802 pKa = 10.52QQSSSCGSSNNPSSSTGYY820 pKa = 10.79VGDD823 pKa = 3.8NTKK826 pKa = 9.6TEE828 pKa = 4.33PLPTTPSPSDD838 pKa = 3.21AASTKK843 pKa = 10.75SVDD846 pKa = 3.41NDD848 pKa = 3.7ALSTPPNGNVDD859 pKa = 3.45DD860 pKa = 4.96HH861 pKa = 5.72QAKK864 pKa = 10.41SNPSSVFDD872 pKa = 4.08FTASCQSSKK881 pKa = 11.02SDD883 pKa = 4.13SGLCTPTASTTSDD896 pKa = 3.23PNQALFYY903 pKa = 10.28QQFSFIKK910 pKa = 9.89EE911 pKa = 4.15YY912 pKa = 11.18NCTPEE917 pKa = 3.88NLRR920 pKa = 11.84LLLEE924 pKa = 4.23TVRR927 pKa = 11.84ARR929 pKa = 11.84LIPMHH934 pKa = 6.64LVHH937 pKa = 7.08PEE939 pKa = 3.77VVPNLDD945 pKa = 3.75SVCEE949 pKa = 3.96PRR951 pKa = 11.84NLSLAFGLSTLCYY964 pKa = 10.27DD965 pKa = 4.19PNPNTGCC972 pKa = 3.92
Molecular weight: 106.41 kDa
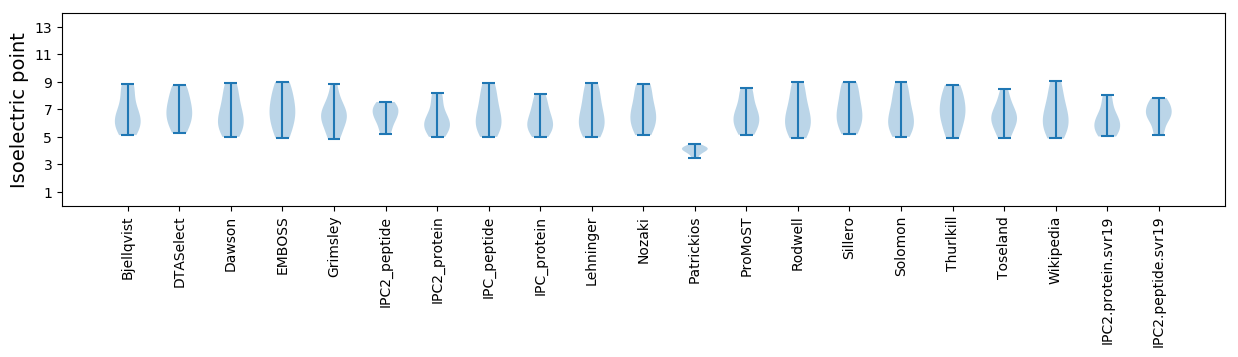
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q83865|Q83865_9REOV 33KD putative nonstructural protein OS=Nilaparvata lugens reovirus OX=33724 PE=4 SV=1
MM1 pKa = 7.36EE2 pKa = 5.3HH3 pKa = 7.26LEE5 pKa = 4.6TITDD9 pKa = 5.05LIFPIFINRR18 pKa = 11.84FRR20 pKa = 11.84QNKK23 pKa = 8.22YY24 pKa = 9.32NNRR27 pKa = 11.84SNTKK31 pKa = 9.89LLNKK35 pKa = 9.44PSPVDD40 pKa = 3.0IKK42 pKa = 10.71RR43 pKa = 11.84YY44 pKa = 7.49YY45 pKa = 10.47SSYY48 pKa = 9.95NSQHH52 pKa = 6.1AVLLRR57 pKa = 11.84VTYY60 pKa = 8.97MPHH63 pKa = 6.27HH64 pKa = 6.22EE65 pKa = 5.19RR66 pKa = 11.84YY67 pKa = 9.2LQSLNIQDD75 pKa = 3.65AQKK78 pKa = 10.68RR79 pKa = 11.84GYY81 pKa = 6.93TAKK84 pKa = 10.72NLDD87 pKa = 3.87LMWYY91 pKa = 10.02RR92 pKa = 11.84LFTGTQRR99 pKa = 11.84GIYY102 pKa = 8.89YY103 pKa = 9.25PPHH106 pKa = 5.87VFNNITKK113 pKa = 10.08KK114 pKa = 10.9YY115 pKa = 9.52GFGLIMNDD123 pKa = 2.59IQTKK127 pKa = 10.35VDD129 pKa = 4.01LEE131 pKa = 4.63TKK133 pKa = 7.49NTHH136 pKa = 5.15YY137 pKa = 11.07VVVDD141 pKa = 3.87FDD143 pKa = 3.88EE144 pKa = 6.74AEE146 pKa = 4.01EE147 pKa = 4.2SDD149 pKa = 5.04NGIQNPGTIIQFSGNGIPMKK169 pKa = 10.63YY170 pKa = 9.87KK171 pKa = 10.74SSTNKK176 pKa = 9.55QFLNNIIKK184 pKa = 10.56LEE186 pKa = 3.95RR187 pKa = 11.84KK188 pKa = 9.42ILDD191 pKa = 3.14EE192 pKa = 4.27SKK194 pKa = 10.8RR195 pKa = 11.84VDD197 pKa = 3.71PLALLNEE204 pKa = 5.04SIQTEE209 pKa = 4.33CAKK212 pKa = 11.04GNVVKK217 pKa = 10.41LQKK220 pKa = 10.49RR221 pKa = 11.84EE222 pKa = 4.34DD223 pKa = 3.11ISFAFEE229 pKa = 3.98CYY231 pKa = 9.67EE232 pKa = 3.8YY233 pKa = 10.55TGNYY237 pKa = 8.59VKK239 pKa = 10.82GNSFIKK245 pKa = 10.16MLLNVDD251 pKa = 4.53FTPIYY256 pKa = 7.93YY257 pKa = 9.88QYY259 pKa = 11.55YY260 pKa = 8.39LTDD263 pKa = 3.36TTNEE267 pKa = 3.78IEE269 pKa = 4.35SYY271 pKa = 9.37PVYY274 pKa = 10.38VQSVLNRR281 pKa = 11.84MKK283 pKa = 10.64LVRR286 pKa = 11.84VPKK289 pKa = 10.25IEE291 pKa = 3.86NEE293 pKa = 3.72IMNGVMPTEE302 pKa = 4.36IAISTCGIVNVRR314 pKa = 11.84FNGWVFTNLAPRR326 pKa = 11.84SKK328 pKa = 10.11FRR330 pKa = 11.84LYY332 pKa = 9.04GTEE335 pKa = 4.07KK336 pKa = 9.93IKK338 pKa = 10.85HH339 pKa = 6.17FIMNYY344 pKa = 8.09VSSFSEE350 pKa = 4.23YY351 pKa = 10.71QMVPYY356 pKa = 10.05FEE358 pKa = 6.23SGAEE362 pKa = 3.67YY363 pKa = 10.37TMRR366 pKa = 11.84IGRR369 pKa = 11.84PIATSFSTFSRR380 pKa = 11.84EE381 pKa = 3.59RR382 pKa = 11.84MAEE385 pKa = 3.87NFKK388 pKa = 10.99DD389 pKa = 3.79AVRR392 pKa = 11.84EE393 pKa = 3.85RR394 pKa = 11.84MPIWVVANKK403 pKa = 10.55GSGKK407 pKa = 8.23TVLRR411 pKa = 11.84KK412 pKa = 9.4EE413 pKa = 4.02LEE415 pKa = 4.19EE416 pKa = 4.41IGFNVIDD423 pKa = 3.68SDD425 pKa = 4.09AYY427 pKa = 8.96GWFVSKK433 pKa = 10.41VAYY436 pKa = 9.02IHH438 pKa = 7.27KK439 pKa = 9.88EE440 pKa = 3.97VNQNTLSTIEE450 pKa = 4.6LSQDD454 pKa = 3.29HH455 pKa = 6.91INRR458 pKa = 11.84LVSEE462 pKa = 4.28VLEE465 pKa = 4.07EE466 pKa = 4.3DD467 pKa = 3.7RR468 pKa = 11.84GISYY472 pKa = 10.13FNVIMYY478 pKa = 10.6SLTKK482 pKa = 9.8QKK484 pKa = 10.9RR485 pKa = 11.84DD486 pKa = 3.16INKK489 pKa = 8.8VLTFPYY495 pKa = 9.06VTHH498 pKa = 6.96RR499 pKa = 11.84WNDD502 pKa = 3.17TLTTFGKK509 pKa = 10.4VFTEE513 pKa = 4.29IADD516 pKa = 3.89SPAIGYY522 pKa = 8.21PRR524 pKa = 11.84FIEE527 pKa = 4.58GCMEE531 pKa = 3.89YY532 pKa = 10.72FRR534 pKa = 11.84VRR536 pKa = 11.84EE537 pKa = 4.08TLHH540 pKa = 6.76KK541 pKa = 10.17PIIFFAHH548 pKa = 6.11TEE550 pKa = 4.22SEE552 pKa = 4.1LSRR555 pKa = 11.84ISIRR559 pKa = 11.84HH560 pKa = 5.27FSCVIYY566 pKa = 10.86NSIDD570 pKa = 3.28SRR572 pKa = 11.84SILSKK577 pKa = 10.41RR578 pKa = 11.84ADD580 pKa = 3.39PVVEE584 pKa = 4.43LMLHH588 pKa = 6.62AYY590 pKa = 10.68YY591 pKa = 11.34YY592 pKa = 8.74MNQVAIDD599 pKa = 4.65KK600 pKa = 10.71IPFSLFRR607 pKa = 11.84QWLGMSNKK615 pKa = 9.26VVDD618 pKa = 4.54EE619 pKa = 4.42SVVAQLIVRR628 pKa = 11.84EE629 pKa = 4.23
MM1 pKa = 7.36EE2 pKa = 5.3HH3 pKa = 7.26LEE5 pKa = 4.6TITDD9 pKa = 5.05LIFPIFINRR18 pKa = 11.84FRR20 pKa = 11.84QNKK23 pKa = 8.22YY24 pKa = 9.32NNRR27 pKa = 11.84SNTKK31 pKa = 9.89LLNKK35 pKa = 9.44PSPVDD40 pKa = 3.0IKK42 pKa = 10.71RR43 pKa = 11.84YY44 pKa = 7.49YY45 pKa = 10.47SSYY48 pKa = 9.95NSQHH52 pKa = 6.1AVLLRR57 pKa = 11.84VTYY60 pKa = 8.97MPHH63 pKa = 6.27HH64 pKa = 6.22EE65 pKa = 5.19RR66 pKa = 11.84YY67 pKa = 9.2LQSLNIQDD75 pKa = 3.65AQKK78 pKa = 10.68RR79 pKa = 11.84GYY81 pKa = 6.93TAKK84 pKa = 10.72NLDD87 pKa = 3.87LMWYY91 pKa = 10.02RR92 pKa = 11.84LFTGTQRR99 pKa = 11.84GIYY102 pKa = 8.89YY103 pKa = 9.25PPHH106 pKa = 5.87VFNNITKK113 pKa = 10.08KK114 pKa = 10.9YY115 pKa = 9.52GFGLIMNDD123 pKa = 2.59IQTKK127 pKa = 10.35VDD129 pKa = 4.01LEE131 pKa = 4.63TKK133 pKa = 7.49NTHH136 pKa = 5.15YY137 pKa = 11.07VVVDD141 pKa = 3.87FDD143 pKa = 3.88EE144 pKa = 6.74AEE146 pKa = 4.01EE147 pKa = 4.2SDD149 pKa = 5.04NGIQNPGTIIQFSGNGIPMKK169 pKa = 10.63YY170 pKa = 9.87KK171 pKa = 10.74SSTNKK176 pKa = 9.55QFLNNIIKK184 pKa = 10.56LEE186 pKa = 3.95RR187 pKa = 11.84KK188 pKa = 9.42ILDD191 pKa = 3.14EE192 pKa = 4.27SKK194 pKa = 10.8RR195 pKa = 11.84VDD197 pKa = 3.71PLALLNEE204 pKa = 5.04SIQTEE209 pKa = 4.33CAKK212 pKa = 11.04GNVVKK217 pKa = 10.41LQKK220 pKa = 10.49RR221 pKa = 11.84EE222 pKa = 4.34DD223 pKa = 3.11ISFAFEE229 pKa = 3.98CYY231 pKa = 9.67EE232 pKa = 3.8YY233 pKa = 10.55TGNYY237 pKa = 8.59VKK239 pKa = 10.82GNSFIKK245 pKa = 10.16MLLNVDD251 pKa = 4.53FTPIYY256 pKa = 7.93YY257 pKa = 9.88QYY259 pKa = 11.55YY260 pKa = 8.39LTDD263 pKa = 3.36TTNEE267 pKa = 3.78IEE269 pKa = 4.35SYY271 pKa = 9.37PVYY274 pKa = 10.38VQSVLNRR281 pKa = 11.84MKK283 pKa = 10.64LVRR286 pKa = 11.84VPKK289 pKa = 10.25IEE291 pKa = 3.86NEE293 pKa = 3.72IMNGVMPTEE302 pKa = 4.36IAISTCGIVNVRR314 pKa = 11.84FNGWVFTNLAPRR326 pKa = 11.84SKK328 pKa = 10.11FRR330 pKa = 11.84LYY332 pKa = 9.04GTEE335 pKa = 4.07KK336 pKa = 9.93IKK338 pKa = 10.85HH339 pKa = 6.17FIMNYY344 pKa = 8.09VSSFSEE350 pKa = 4.23YY351 pKa = 10.71QMVPYY356 pKa = 10.05FEE358 pKa = 6.23SGAEE362 pKa = 3.67YY363 pKa = 10.37TMRR366 pKa = 11.84IGRR369 pKa = 11.84PIATSFSTFSRR380 pKa = 11.84EE381 pKa = 3.59RR382 pKa = 11.84MAEE385 pKa = 3.87NFKK388 pKa = 10.99DD389 pKa = 3.79AVRR392 pKa = 11.84EE393 pKa = 3.85RR394 pKa = 11.84MPIWVVANKK403 pKa = 10.55GSGKK407 pKa = 8.23TVLRR411 pKa = 11.84KK412 pKa = 9.4EE413 pKa = 4.02LEE415 pKa = 4.19EE416 pKa = 4.41IGFNVIDD423 pKa = 3.68SDD425 pKa = 4.09AYY427 pKa = 8.96GWFVSKK433 pKa = 10.41VAYY436 pKa = 9.02IHH438 pKa = 7.27KK439 pKa = 9.88EE440 pKa = 3.97VNQNTLSTIEE450 pKa = 4.6LSQDD454 pKa = 3.29HH455 pKa = 6.91INRR458 pKa = 11.84LVSEE462 pKa = 4.28VLEE465 pKa = 4.07EE466 pKa = 4.3DD467 pKa = 3.7RR468 pKa = 11.84GISYY472 pKa = 10.13FNVIMYY478 pKa = 10.6SLTKK482 pKa = 9.8QKK484 pKa = 10.9RR485 pKa = 11.84DD486 pKa = 3.16INKK489 pKa = 8.8VLTFPYY495 pKa = 9.06VTHH498 pKa = 6.96RR499 pKa = 11.84WNDD502 pKa = 3.17TLTTFGKK509 pKa = 10.4VFTEE513 pKa = 4.29IADD516 pKa = 3.89SPAIGYY522 pKa = 8.21PRR524 pKa = 11.84FIEE527 pKa = 4.58GCMEE531 pKa = 3.89YY532 pKa = 10.72FRR534 pKa = 11.84VRR536 pKa = 11.84EE537 pKa = 4.08TLHH540 pKa = 6.76KK541 pKa = 10.17PIIFFAHH548 pKa = 6.11TEE550 pKa = 4.22SEE552 pKa = 4.1LSRR555 pKa = 11.84ISIRR559 pKa = 11.84HH560 pKa = 5.27FSCVIYY566 pKa = 10.86NSIDD570 pKa = 3.28SRR572 pKa = 11.84SILSKK577 pKa = 10.41RR578 pKa = 11.84ADD580 pKa = 3.39PVVEE584 pKa = 4.43LMLHH588 pKa = 6.62AYY590 pKa = 10.68YY591 pKa = 11.34YY592 pKa = 8.74MNQVAIDD599 pKa = 4.65KK600 pKa = 10.71IPFSLFRR607 pKa = 11.84QWLGMSNKK615 pKa = 9.26VVDD618 pKa = 4.54EE619 pKa = 4.42SVVAQLIVRR628 pKa = 11.84EE629 pKa = 4.23
Molecular weight: 73.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8916 |
206 |
1442 |
810.5 |
92.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.957 ± 0.293 | 1.447 ± 0.196 |
6.18 ± 0.302 | 5.002 ± 0.366 |
5.316 ± 0.239 | 4.565 ± 0.25 |
2.131 ± 0.146 | 7.279 ± 0.511 |
6.292 ± 0.262 | 8.636 ± 0.408 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.838 ± 0.099 | 6.819 ± 0.239 |
3.892 ± 0.312 | 3.297 ± 0.174 |
4.307 ± 0.339 | 8.457 ± 0.523 |
6.886 ± 0.242 | 6.359 ± 0.251 |
0.729 ± 0.073 | 4.61 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
