
Fusibacter sp. A1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Eubacteriales Family XII. Incertae Sedis; Fusibacter; unclassified Fusibacter
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
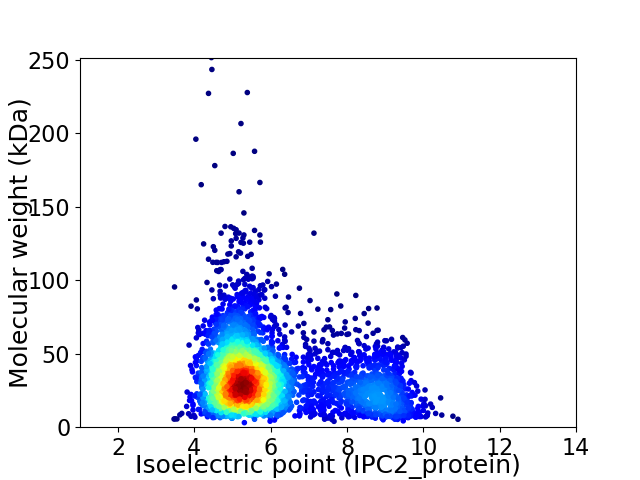
Virtual 2D-PAGE plot for 3729 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q1ZTF1|A0A4Q1ZTF1_9FIRM Ribonuclease PH OS=Fusibacter sp. A1 OX=2283630 GN=rph PE=3 SV=1
MM1 pKa = 7.31KK2 pKa = 10.01RR3 pKa = 11.84HH4 pKa = 5.56FKK6 pKa = 9.87VRR8 pKa = 11.84HH9 pKa = 4.76VFSFVLILALVMAGVACQKK28 pKa = 10.34QLSAKK33 pKa = 8.02EE34 pKa = 4.17TVEE37 pKa = 3.83RR38 pKa = 11.84LYY40 pKa = 11.08EE41 pKa = 3.92EE42 pKa = 4.55SLNLDD47 pKa = 3.48AYY49 pKa = 9.35SAKK52 pKa = 10.6FEE54 pKa = 4.79GDD56 pKa = 2.77MSMEE60 pKa = 5.1FIGEE64 pKa = 4.22TDD66 pKa = 3.45PMIEE70 pKa = 4.09QYY72 pKa = 11.47LGMFKK77 pKa = 10.31NIKK80 pKa = 9.43MSGDD84 pKa = 3.48MQYY87 pKa = 11.48KK88 pKa = 8.83DD89 pKa = 3.24TDD91 pKa = 4.16RR92 pKa = 11.84IGDD95 pKa = 3.49YY96 pKa = 11.54AMNYY100 pKa = 7.98TLDD103 pKa = 3.75MNGMSMAIEE112 pKa = 4.98FYY114 pKa = 10.86FDD116 pKa = 3.61GSRR119 pKa = 11.84MIVNYY124 pKa = 9.49PLLPNYY130 pKa = 9.69IVIDD134 pKa = 4.13LPEE137 pKa = 4.79LLGMLNEE144 pKa = 4.1EE145 pKa = 3.97QEE147 pKa = 4.53APVEE151 pKa = 3.84LDD153 pKa = 3.31YY154 pKa = 11.69EE155 pKa = 4.94SIVQDD160 pKa = 4.09LEE162 pKa = 4.19ALTSEE167 pKa = 4.75FMPKK171 pKa = 10.45LSGEE175 pKa = 4.12IIKK178 pKa = 11.0LIDD181 pKa = 3.78EE182 pKa = 4.74NSLEE186 pKa = 4.39LLDD189 pKa = 5.23EE190 pKa = 4.33YY191 pKa = 11.16TFTVDD196 pKa = 4.05GEE198 pKa = 4.92SVTSKK203 pKa = 10.61AVKK206 pKa = 10.53VNLDD210 pKa = 3.47TDD212 pKa = 3.94LMIGMMNSLFEE223 pKa = 3.86QAKK226 pKa = 9.07NSEE229 pKa = 4.2VVYY232 pKa = 10.19QIVKK236 pKa = 10.42KK237 pKa = 10.85YY238 pKa = 10.92DD239 pKa = 3.09IDD241 pKa = 4.71DD242 pKa = 3.88EE243 pKa = 4.68VGSFEE248 pKa = 5.75DD249 pKa = 3.86YY250 pKa = 10.67QAGFDD255 pKa = 3.67GLKK258 pKa = 10.63ASFEE262 pKa = 4.32EE263 pKa = 4.31EE264 pKa = 4.04MNLSEE269 pKa = 5.16FEE271 pKa = 4.28EE272 pKa = 4.26MMKK275 pKa = 10.58NISYY279 pKa = 8.96TYY281 pKa = 10.45ILGYY285 pKa = 10.3DD286 pKa = 2.83KK287 pKa = 11.02NYY289 pKa = 10.4RR290 pKa = 11.84VNHH293 pKa = 5.7MALDD297 pKa = 4.02LTMKK301 pKa = 10.13IDD303 pKa = 4.1DD304 pKa = 4.18EE305 pKa = 5.18SIGTEE310 pKa = 3.38IEE312 pKa = 3.98MTMGAVYY319 pKa = 10.92AMNYY323 pKa = 8.08DD324 pKa = 3.32AKK326 pKa = 10.59EE327 pKa = 3.82IEE329 pKa = 4.49FPEE332 pKa = 4.33LTDD335 pKa = 3.78EE336 pKa = 4.2NSMSIMEE343 pKa = 5.12LFPDD347 pKa = 3.93LSGYY351 pKa = 10.66GSYY354 pKa = 11.05EE355 pKa = 4.03DD356 pKa = 3.87GGDD359 pKa = 3.62YY360 pKa = 10.95EE361 pKa = 6.14DD362 pKa = 4.48YY363 pKa = 11.5VPVNYY368 pKa = 10.24FIEE371 pKa = 4.5EE372 pKa = 4.03QVLMLDD378 pKa = 6.11DD379 pKa = 4.22IAQDD383 pKa = 3.58LTLLTEE389 pKa = 3.99TTEE392 pKa = 3.81IEE394 pKa = 4.44EE395 pKa = 4.24VFEE398 pKa = 4.61SYY400 pKa = 11.17VYY402 pKa = 10.73DD403 pKa = 3.69SYY405 pKa = 11.88FEE407 pKa = 4.15VMNLYY412 pKa = 10.2YY413 pKa = 9.92ATSEE417 pKa = 4.44GEE419 pKa = 4.07MILYY423 pKa = 8.73PNQEE427 pKa = 4.2LPDD430 pKa = 3.85NYY432 pKa = 10.78DD433 pKa = 3.01PRR435 pKa = 11.84EE436 pKa = 4.08RR437 pKa = 11.84PWYY440 pKa = 9.62TEE442 pKa = 3.75ALLQGTMIGEE452 pKa = 4.73LYY454 pKa = 10.42EE455 pKa = 4.57DD456 pKa = 3.86AASGRR461 pKa = 11.84FVQSISKK468 pKa = 9.03TIVLDD473 pKa = 3.8GEE475 pKa = 4.69VIGVVGIDD483 pKa = 3.3IFVDD487 pKa = 3.67
MM1 pKa = 7.31KK2 pKa = 10.01RR3 pKa = 11.84HH4 pKa = 5.56FKK6 pKa = 9.87VRR8 pKa = 11.84HH9 pKa = 4.76VFSFVLILALVMAGVACQKK28 pKa = 10.34QLSAKK33 pKa = 8.02EE34 pKa = 4.17TVEE37 pKa = 3.83RR38 pKa = 11.84LYY40 pKa = 11.08EE41 pKa = 3.92EE42 pKa = 4.55SLNLDD47 pKa = 3.48AYY49 pKa = 9.35SAKK52 pKa = 10.6FEE54 pKa = 4.79GDD56 pKa = 2.77MSMEE60 pKa = 5.1FIGEE64 pKa = 4.22TDD66 pKa = 3.45PMIEE70 pKa = 4.09QYY72 pKa = 11.47LGMFKK77 pKa = 10.31NIKK80 pKa = 9.43MSGDD84 pKa = 3.48MQYY87 pKa = 11.48KK88 pKa = 8.83DD89 pKa = 3.24TDD91 pKa = 4.16RR92 pKa = 11.84IGDD95 pKa = 3.49YY96 pKa = 11.54AMNYY100 pKa = 7.98TLDD103 pKa = 3.75MNGMSMAIEE112 pKa = 4.98FYY114 pKa = 10.86FDD116 pKa = 3.61GSRR119 pKa = 11.84MIVNYY124 pKa = 9.49PLLPNYY130 pKa = 9.69IVIDD134 pKa = 4.13LPEE137 pKa = 4.79LLGMLNEE144 pKa = 4.1EE145 pKa = 3.97QEE147 pKa = 4.53APVEE151 pKa = 3.84LDD153 pKa = 3.31YY154 pKa = 11.69EE155 pKa = 4.94SIVQDD160 pKa = 4.09LEE162 pKa = 4.19ALTSEE167 pKa = 4.75FMPKK171 pKa = 10.45LSGEE175 pKa = 4.12IIKK178 pKa = 11.0LIDD181 pKa = 3.78EE182 pKa = 4.74NSLEE186 pKa = 4.39LLDD189 pKa = 5.23EE190 pKa = 4.33YY191 pKa = 11.16TFTVDD196 pKa = 4.05GEE198 pKa = 4.92SVTSKK203 pKa = 10.61AVKK206 pKa = 10.53VNLDD210 pKa = 3.47TDD212 pKa = 3.94LMIGMMNSLFEE223 pKa = 3.86QAKK226 pKa = 9.07NSEE229 pKa = 4.2VVYY232 pKa = 10.19QIVKK236 pKa = 10.42KK237 pKa = 10.85YY238 pKa = 10.92DD239 pKa = 3.09IDD241 pKa = 4.71DD242 pKa = 3.88EE243 pKa = 4.68VGSFEE248 pKa = 5.75DD249 pKa = 3.86YY250 pKa = 10.67QAGFDD255 pKa = 3.67GLKK258 pKa = 10.63ASFEE262 pKa = 4.32EE263 pKa = 4.31EE264 pKa = 4.04MNLSEE269 pKa = 5.16FEE271 pKa = 4.28EE272 pKa = 4.26MMKK275 pKa = 10.58NISYY279 pKa = 8.96TYY281 pKa = 10.45ILGYY285 pKa = 10.3DD286 pKa = 2.83KK287 pKa = 11.02NYY289 pKa = 10.4RR290 pKa = 11.84VNHH293 pKa = 5.7MALDD297 pKa = 4.02LTMKK301 pKa = 10.13IDD303 pKa = 4.1DD304 pKa = 4.18EE305 pKa = 5.18SIGTEE310 pKa = 3.38IEE312 pKa = 3.98MTMGAVYY319 pKa = 10.92AMNYY323 pKa = 8.08DD324 pKa = 3.32AKK326 pKa = 10.59EE327 pKa = 3.82IEE329 pKa = 4.49FPEE332 pKa = 4.33LTDD335 pKa = 3.78EE336 pKa = 4.2NSMSIMEE343 pKa = 5.12LFPDD347 pKa = 3.93LSGYY351 pKa = 10.66GSYY354 pKa = 11.05EE355 pKa = 4.03DD356 pKa = 3.87GGDD359 pKa = 3.62YY360 pKa = 10.95EE361 pKa = 6.14DD362 pKa = 4.48YY363 pKa = 11.5VPVNYY368 pKa = 10.24FIEE371 pKa = 4.5EE372 pKa = 4.03QVLMLDD378 pKa = 6.11DD379 pKa = 4.22IAQDD383 pKa = 3.58LTLLTEE389 pKa = 3.99TTEE392 pKa = 3.81IEE394 pKa = 4.44EE395 pKa = 4.24VFEE398 pKa = 4.61SYY400 pKa = 11.17VYY402 pKa = 10.73DD403 pKa = 3.69SYY405 pKa = 11.88FEE407 pKa = 4.15VMNLYY412 pKa = 10.2YY413 pKa = 9.92ATSEE417 pKa = 4.44GEE419 pKa = 4.07MILYY423 pKa = 8.73PNQEE427 pKa = 4.2LPDD430 pKa = 3.85NYY432 pKa = 10.78DD433 pKa = 3.01PRR435 pKa = 11.84EE436 pKa = 4.08RR437 pKa = 11.84PWYY440 pKa = 9.62TEE442 pKa = 3.75ALLQGTMIGEE452 pKa = 4.73LYY454 pKa = 10.42EE455 pKa = 4.57DD456 pKa = 3.86AASGRR461 pKa = 11.84FVQSISKK468 pKa = 9.03TIVLDD473 pKa = 3.8GEE475 pKa = 4.69VIGVVGIDD483 pKa = 3.3IFVDD487 pKa = 3.67
Molecular weight: 55.9 kDa
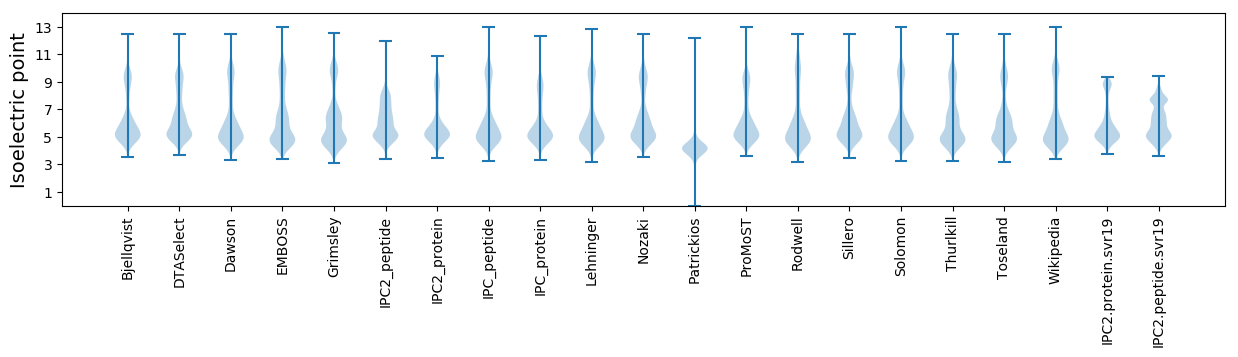
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q1ZTC1|A0A4Q1ZTC1_9FIRM DUF1846 domain-containing protein OS=Fusibacter sp. A1 OX=2283630 GN=DWB64_14555 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 8.57KK9 pKa = 7.61RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.25RR14 pKa = 11.84EE15 pKa = 3.66HH16 pKa = 6.26GFRR19 pKa = 11.84KK20 pKa = 9.93RR21 pKa = 11.84MSSSAGRR28 pKa = 11.84NVLKK32 pKa = 10.44RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.5LSAA44 pKa = 3.94
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 8.57KK9 pKa = 7.61RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.25RR14 pKa = 11.84EE15 pKa = 3.66HH16 pKa = 6.26GFRR19 pKa = 11.84KK20 pKa = 9.93RR21 pKa = 11.84MSSSAGRR28 pKa = 11.84NVLKK32 pKa = 10.44RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.5LSAA44 pKa = 3.94
Molecular weight: 5.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1207282 |
31 |
2347 |
323.8 |
36.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.73 ± 0.042 | 0.889 ± 0.015 |
5.958 ± 0.032 | 7.175 ± 0.04 |
4.382 ± 0.029 | 6.616 ± 0.041 |
2.123 ± 0.02 | 8.07 ± 0.035 |
6.84 ± 0.03 | 9.934 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.076 ± 0.019 | 4.331 ± 0.024 |
3.025 ± 0.023 | 2.996 ± 0.022 |
3.991 ± 0.026 | 6.401 ± 0.032 |
5.474 ± 0.034 | 7.317 ± 0.026 |
0.806 ± 0.015 | 3.866 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
