
Anopheles sinensis (Mosquito)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Diptera; Nematocera; Culicomorpha; Culicoidea; Culicidae;
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
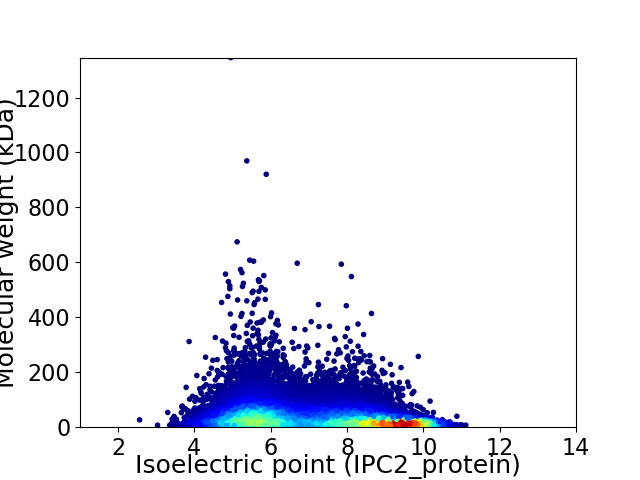
Virtual 2D-PAGE plot for 19343 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
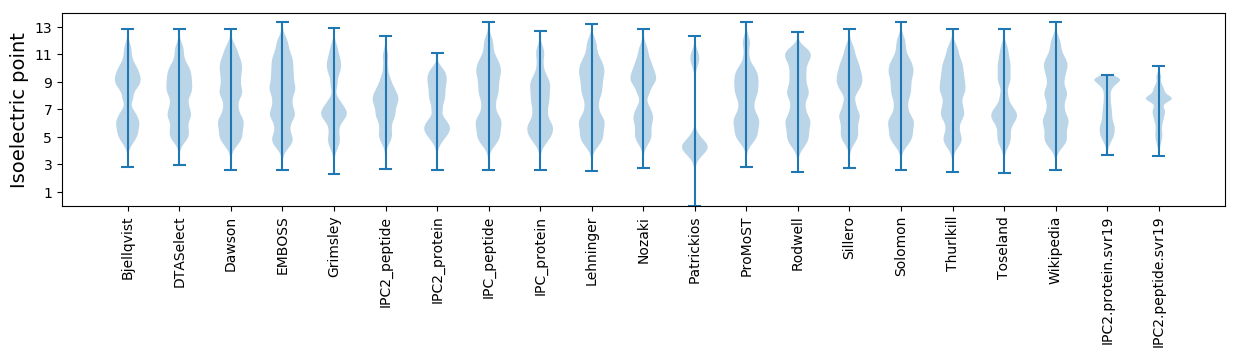
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A084W8K2|A0A084W8K2_ANOSI Biotin-protein ligase OS=Anopheles sinensis OX=74873 GN=ZHAS_00014545 PE=4 SV=1
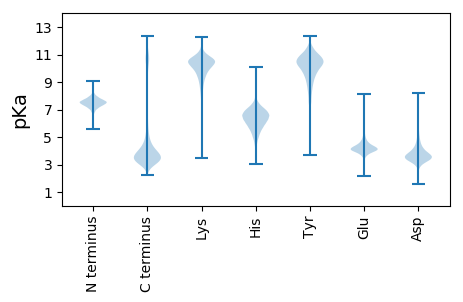
MM1 pKa = 7.38RR2 pKa = 11.84TFGFAVATVLCAIQLTAALPRR23 pKa = 11.84PDD25 pKa = 4.38FGLSIDD31 pKa = 4.27VYY33 pKa = 11.11SADD36 pKa = 3.69KK37 pKa = 10.81TSDD40 pKa = 3.32VADD43 pKa = 3.88AANNDD48 pKa = 4.24LVTLWSTSTITLNANYY64 pKa = 7.7DD65 pKa = 3.82TLGEE69 pKa = 4.06ILTEE73 pKa = 3.79ITAIGTDD80 pKa = 3.13ASANIKK86 pKa = 10.43AVFDD90 pKa = 4.88AIIDD94 pKa = 4.04LADD97 pKa = 3.6ATTDD101 pKa = 3.17PTAAFDD107 pKa = 3.89AAIDD111 pKa = 3.93AVDD114 pKa = 4.0ALITYY119 pKa = 7.73ITSEE123 pKa = 4.38TVGLSVNYY131 pKa = 10.46GNLDD135 pKa = 3.51TALSTNEE142 pKa = 3.79IVVQLEE148 pKa = 4.07DD149 pKa = 4.16AFGEE153 pKa = 4.29LADD156 pKa = 3.9GLARR160 pKa = 11.84LGAALTALKK169 pKa = 10.36AQVLAAVTTAGSGTVTKK186 pKa = 10.73AILRR190 pKa = 11.84QKK192 pKa = 10.93LSIADD197 pKa = 3.82TSDD200 pKa = 3.14LTVSVVDD207 pKa = 4.06VSSSLQLIFFILEE220 pKa = 3.66QSKK223 pKa = 10.44EE224 pKa = 3.95FLQLADD230 pKa = 4.27AYY232 pKa = 9.68IISSGVAGQDD242 pKa = 2.85AVDD245 pKa = 4.29YY246 pKa = 10.9INEE249 pKa = 3.96QLQVLSGEE257 pKa = 4.23VEE259 pKa = 4.4SYY261 pKa = 11.2KK262 pKa = 10.92DD263 pKa = 3.19AADD266 pKa = 4.16TIKK269 pKa = 8.15TTVTDD274 pKa = 3.83TFDD277 pKa = 3.32GSTDD281 pKa = 3.11LGARR285 pKa = 11.84DD286 pKa = 4.22LTPIAAIKK294 pKa = 9.72TEE296 pKa = 3.68VDD298 pKa = 3.06ATVTTFGANIDD309 pKa = 3.94TAVTAIEE316 pKa = 4.18ASFTGYY322 pKa = 9.97QADD325 pKa = 3.5ILLVSTGYY333 pKa = 9.17ATFYY337 pKa = 10.96SDD339 pKa = 3.71NACGHH344 pKa = 6.35IFDD347 pKa = 5.46LVTVLINQGPYY358 pKa = 10.54AEE360 pKa = 4.34YY361 pKa = 10.37CYY363 pKa = 10.79EE364 pKa = 4.15KK365 pKa = 11.26YY366 pKa = 11.02GDD368 pKa = 3.65AALALFDD375 pKa = 4.39SNARR379 pKa = 11.84VAGEE383 pKa = 4.12CVDD386 pKa = 4.57RR387 pKa = 11.84EE388 pKa = 4.18ITRR391 pKa = 11.84LLTLQDD397 pKa = 3.62VLISIAEE404 pKa = 3.94QIAFSVEE411 pKa = 3.9DD412 pKa = 4.27LLATLDD418 pKa = 3.68FCLNSVTEE426 pKa = 4.77CNDD429 pKa = 2.88SDD431 pKa = 3.49IDD433 pKa = 3.82MYY435 pKa = 11.29LGAIHH440 pKa = 7.13AEE442 pKa = 3.69IDD444 pKa = 3.48ASHH447 pKa = 6.97LEE449 pKa = 4.09VLTDD453 pKa = 4.3LVDD456 pKa = 4.95DD457 pKa = 4.19EE458 pKa = 4.9TDD460 pKa = 2.93AGLARR465 pKa = 11.84LQACFVAARR474 pKa = 11.84YY475 pKa = 9.46DD476 pKa = 3.35ILRR479 pKa = 11.84EE480 pKa = 3.82IDD482 pKa = 4.62GMTTDD487 pKa = 4.3IAACEE492 pKa = 4.3TNGPNPPSPRR502 pKa = 3.46
MM1 pKa = 7.38RR2 pKa = 11.84TFGFAVATVLCAIQLTAALPRR23 pKa = 11.84PDD25 pKa = 4.38FGLSIDD31 pKa = 4.27VYY33 pKa = 11.11SADD36 pKa = 3.69KK37 pKa = 10.81TSDD40 pKa = 3.32VADD43 pKa = 3.88AANNDD48 pKa = 4.24LVTLWSTSTITLNANYY64 pKa = 7.7DD65 pKa = 3.82TLGEE69 pKa = 4.06ILTEE73 pKa = 3.79ITAIGTDD80 pKa = 3.13ASANIKK86 pKa = 10.43AVFDD90 pKa = 4.88AIIDD94 pKa = 4.04LADD97 pKa = 3.6ATTDD101 pKa = 3.17PTAAFDD107 pKa = 3.89AAIDD111 pKa = 3.93AVDD114 pKa = 4.0ALITYY119 pKa = 7.73ITSEE123 pKa = 4.38TVGLSVNYY131 pKa = 10.46GNLDD135 pKa = 3.51TALSTNEE142 pKa = 3.79IVVQLEE148 pKa = 4.07DD149 pKa = 4.16AFGEE153 pKa = 4.29LADD156 pKa = 3.9GLARR160 pKa = 11.84LGAALTALKK169 pKa = 10.36AQVLAAVTTAGSGTVTKK186 pKa = 10.73AILRR190 pKa = 11.84QKK192 pKa = 10.93LSIADD197 pKa = 3.82TSDD200 pKa = 3.14LTVSVVDD207 pKa = 4.06VSSSLQLIFFILEE220 pKa = 3.66QSKK223 pKa = 10.44EE224 pKa = 3.95FLQLADD230 pKa = 4.27AYY232 pKa = 9.68IISSGVAGQDD242 pKa = 2.85AVDD245 pKa = 4.29YY246 pKa = 10.9INEE249 pKa = 3.96QLQVLSGEE257 pKa = 4.23VEE259 pKa = 4.4SYY261 pKa = 11.2KK262 pKa = 10.92DD263 pKa = 3.19AADD266 pKa = 4.16TIKK269 pKa = 8.15TTVTDD274 pKa = 3.83TFDD277 pKa = 3.32GSTDD281 pKa = 3.11LGARR285 pKa = 11.84DD286 pKa = 4.22LTPIAAIKK294 pKa = 9.72TEE296 pKa = 3.68VDD298 pKa = 3.06ATVTTFGANIDD309 pKa = 3.94TAVTAIEE316 pKa = 4.18ASFTGYY322 pKa = 9.97QADD325 pKa = 3.5ILLVSTGYY333 pKa = 9.17ATFYY337 pKa = 10.96SDD339 pKa = 3.71NACGHH344 pKa = 6.35IFDD347 pKa = 5.46LVTVLINQGPYY358 pKa = 10.54AEE360 pKa = 4.34YY361 pKa = 10.37CYY363 pKa = 10.79EE364 pKa = 4.15KK365 pKa = 11.26YY366 pKa = 11.02GDD368 pKa = 3.65AALALFDD375 pKa = 4.39SNARR379 pKa = 11.84VAGEE383 pKa = 4.12CVDD386 pKa = 4.57RR387 pKa = 11.84EE388 pKa = 4.18ITRR391 pKa = 11.84LLTLQDD397 pKa = 3.62VLISIAEE404 pKa = 3.94QIAFSVEE411 pKa = 3.9DD412 pKa = 4.27LLATLDD418 pKa = 3.68FCLNSVTEE426 pKa = 4.77CNDD429 pKa = 2.88SDD431 pKa = 3.49IDD433 pKa = 3.82MYY435 pKa = 11.29LGAIHH440 pKa = 7.13AEE442 pKa = 3.69IDD444 pKa = 3.48ASHH447 pKa = 6.97LEE449 pKa = 4.09VLTDD453 pKa = 4.3LVDD456 pKa = 4.95DD457 pKa = 4.19EE458 pKa = 4.9TDD460 pKa = 2.93AGLARR465 pKa = 11.84LQACFVAARR474 pKa = 11.84YY475 pKa = 9.46DD476 pKa = 3.35ILRR479 pKa = 11.84EE480 pKa = 3.82IDD482 pKa = 4.62GMTTDD487 pKa = 4.3IAACEE492 pKa = 4.3TNGPNPPSPRR502 pKa = 3.46
Molecular weight: 53.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A084VFU9|A0A084VFU9_ANOSI Probable U3 small nucleolar RNA-associated protein 11 OS=Anopheles sinensis OX=74873 GN=ZHAS_00004010 PE=3 SV=1
MM1 pKa = 7.16MNAAILIGLGMISSPNSHH19 pKa = 7.23LKK21 pKa = 10.32RR22 pKa = 11.84SRR24 pKa = 11.84NSLPSRR30 pKa = 11.84GLPFLFLPRR39 pKa = 11.84RR40 pKa = 11.84LNKK43 pKa = 9.4QANNGKK49 pKa = 8.31GTVFQGRR56 pKa = 11.84RR57 pKa = 11.84SAPGGKK63 pKa = 9.54GSKK66 pKa = 9.68KK67 pKa = 9.88RR68 pKa = 11.84ALL70 pKa = 3.66
MM1 pKa = 7.16MNAAILIGLGMISSPNSHH19 pKa = 7.23LKK21 pKa = 10.32RR22 pKa = 11.84SRR24 pKa = 11.84NSLPSRR30 pKa = 11.84GLPFLFLPRR39 pKa = 11.84RR40 pKa = 11.84LNKK43 pKa = 9.4QANNGKK49 pKa = 8.31GTVFQGRR56 pKa = 11.84RR57 pKa = 11.84SAPGGKK63 pKa = 9.54GSKK66 pKa = 9.68KK67 pKa = 9.88RR68 pKa = 11.84ALL70 pKa = 3.66
Molecular weight: 7.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7280472 |
47 |
11741 |
376.4 |
41.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.327 ± 0.018 | 1.919 ± 0.017 |
5.214 ± 0.015 | 6.241 ± 0.025 |
3.571 ± 0.015 | 6.893 ± 0.03 |
2.682 ± 0.014 | 4.744 ± 0.018 |
5.248 ± 0.022 | 8.738 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.289 ± 0.009 | 4.403 ± 0.013 |
5.656 ± 0.025 | 4.373 ± 0.018 |
6.076 ± 0.02 | 8.23 ± 0.033 |
6.079 ± 0.02 | 6.341 ± 0.015 |
1.038 ± 0.007 | 2.886 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
