
Circoviridae 11 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
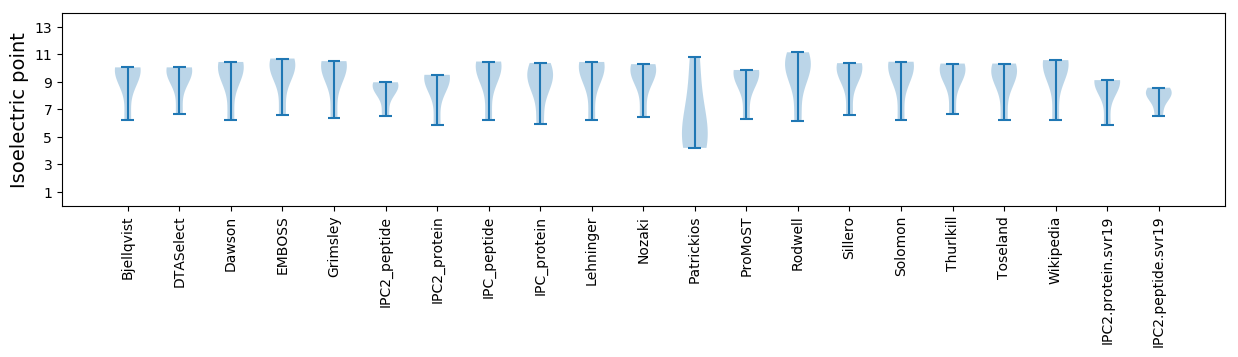
Average proteome isoelectric point is 8.4
Get precalculated fractions of proteins
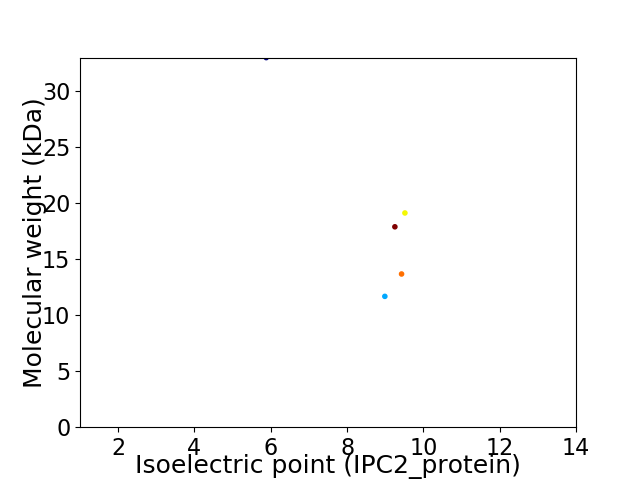
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5TNE1|S5TNE1_9CIRC Uncharacterized protein OS=Circoviridae 11 LDMD-2013 OX=1379715 PE=4 SV=1
MM1 pKa = 6.89PTPRR5 pKa = 11.84GVYY8 pKa = 8.76WCWTLNNYY16 pKa = 7.76TPEE19 pKa = 4.19EE20 pKa = 4.01LSHH23 pKa = 6.9CRR25 pKa = 11.84TLCDD29 pKa = 3.04RR30 pKa = 11.84AEE32 pKa = 4.31DD33 pKa = 3.59VTYY36 pKa = 10.7VCFSRR41 pKa = 11.84EE42 pKa = 4.13VGDD45 pKa = 3.52SGTPHH50 pKa = 5.94LQGYY54 pKa = 9.69LEE56 pKa = 4.43LAAKK60 pKa = 10.08LRR62 pKa = 11.84LGGLRR67 pKa = 11.84KK68 pKa = 9.73LDD70 pKa = 3.64GLGRR74 pKa = 11.84AHH76 pKa = 7.52LEE78 pKa = 4.03LRR80 pKa = 11.84KK81 pKa = 8.78GTQAEE86 pKa = 4.33AIDD89 pKa = 3.97YY90 pKa = 9.52CRR92 pKa = 11.84KK93 pKa = 10.43DD94 pKa = 3.3GNEE97 pKa = 3.82TFEE100 pKa = 4.0EE101 pKa = 4.36HH102 pKa = 5.89GTRR105 pKa = 11.84KK106 pKa = 8.33TSSQGKK112 pKa = 9.89RR113 pKa = 11.84MDD115 pKa = 3.71LEE117 pKa = 4.2AVKK120 pKa = 10.8ALLDD124 pKa = 3.66GGAPLRR130 pKa = 11.84EE131 pKa = 4.2VADD134 pKa = 3.41QHH136 pKa = 6.6FGTFVRR142 pKa = 11.84YY143 pKa = 9.66GRR145 pKa = 11.84GIAQYY150 pKa = 11.18QLVTAKK156 pKa = 10.13KK157 pKa = 8.15RR158 pKa = 11.84ARR160 pKa = 11.84DD161 pKa = 3.67VSVHH165 pKa = 5.34VLWGAPGVGKK175 pKa = 8.12TRR177 pKa = 11.84WIFEE181 pKa = 3.98RR182 pKa = 11.84HH183 pKa = 6.56ADD185 pKa = 4.74DD186 pKa = 5.84LWICNDD192 pKa = 3.73PTLQWFDD199 pKa = 3.6GYY201 pKa = 10.9QGEE204 pKa = 4.69SVVLIDD210 pKa = 5.02DD211 pKa = 4.16FRR213 pKa = 11.84GEE215 pKa = 3.86ASPAFMLRR223 pKa = 11.84LLDD226 pKa = 4.49RR227 pKa = 11.84YY228 pKa = 9.47PLQVPIKK235 pKa = 10.58GGFTEE240 pKa = 4.41WRR242 pKa = 11.84ATTIYY247 pKa = 9.01ITSNVAPPFGMEE259 pKa = 4.66SIVAPLARR267 pKa = 11.84RR268 pKa = 11.84LSIVQEE274 pKa = 4.19LPKK277 pKa = 10.16QEE279 pKa = 3.99YY280 pKa = 10.34DD281 pKa = 3.06ITEE284 pKa = 4.01AALNEE289 pKa = 4.24IFNN292 pKa = 4.4
MM1 pKa = 6.89PTPRR5 pKa = 11.84GVYY8 pKa = 8.76WCWTLNNYY16 pKa = 7.76TPEE19 pKa = 4.19EE20 pKa = 4.01LSHH23 pKa = 6.9CRR25 pKa = 11.84TLCDD29 pKa = 3.04RR30 pKa = 11.84AEE32 pKa = 4.31DD33 pKa = 3.59VTYY36 pKa = 10.7VCFSRR41 pKa = 11.84EE42 pKa = 4.13VGDD45 pKa = 3.52SGTPHH50 pKa = 5.94LQGYY54 pKa = 9.69LEE56 pKa = 4.43LAAKK60 pKa = 10.08LRR62 pKa = 11.84LGGLRR67 pKa = 11.84KK68 pKa = 9.73LDD70 pKa = 3.64GLGRR74 pKa = 11.84AHH76 pKa = 7.52LEE78 pKa = 4.03LRR80 pKa = 11.84KK81 pKa = 8.78GTQAEE86 pKa = 4.33AIDD89 pKa = 3.97YY90 pKa = 9.52CRR92 pKa = 11.84KK93 pKa = 10.43DD94 pKa = 3.3GNEE97 pKa = 3.82TFEE100 pKa = 4.0EE101 pKa = 4.36HH102 pKa = 5.89GTRR105 pKa = 11.84KK106 pKa = 8.33TSSQGKK112 pKa = 9.89RR113 pKa = 11.84MDD115 pKa = 3.71LEE117 pKa = 4.2AVKK120 pKa = 10.8ALLDD124 pKa = 3.66GGAPLRR130 pKa = 11.84EE131 pKa = 4.2VADD134 pKa = 3.41QHH136 pKa = 6.6FGTFVRR142 pKa = 11.84YY143 pKa = 9.66GRR145 pKa = 11.84GIAQYY150 pKa = 11.18QLVTAKK156 pKa = 10.13KK157 pKa = 8.15RR158 pKa = 11.84ARR160 pKa = 11.84DD161 pKa = 3.67VSVHH165 pKa = 5.34VLWGAPGVGKK175 pKa = 8.12TRR177 pKa = 11.84WIFEE181 pKa = 3.98RR182 pKa = 11.84HH183 pKa = 6.56ADD185 pKa = 4.74DD186 pKa = 5.84LWICNDD192 pKa = 3.73PTLQWFDD199 pKa = 3.6GYY201 pKa = 10.9QGEE204 pKa = 4.69SVVLIDD210 pKa = 5.02DD211 pKa = 4.16FRR213 pKa = 11.84GEE215 pKa = 3.86ASPAFMLRR223 pKa = 11.84LLDD226 pKa = 4.49RR227 pKa = 11.84YY228 pKa = 9.47PLQVPIKK235 pKa = 10.58GGFTEE240 pKa = 4.41WRR242 pKa = 11.84ATTIYY247 pKa = 9.01ITSNVAPPFGMEE259 pKa = 4.66SIVAPLARR267 pKa = 11.84RR268 pKa = 11.84LSIVQEE274 pKa = 4.19LPKK277 pKa = 10.16QEE279 pKa = 3.99YY280 pKa = 10.34DD281 pKa = 3.06ITEE284 pKa = 4.01AALNEE289 pKa = 4.24IFNN292 pKa = 4.4
Molecular weight: 33.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5SYE3|S5SYE3_9CIRC Uncharacterized protein OS=Circoviridae 11 LDMD-2013 OX=1379715 PE=4 SV=1
MM1 pKa = 7.49RR2 pKa = 11.84VDD4 pKa = 3.56IPEE7 pKa = 3.83MGYY10 pKa = 10.35NLVSRR15 pKa = 11.84DD16 pKa = 2.93VDD18 pKa = 5.29LIILLYY24 pKa = 10.58ARR26 pKa = 11.84IWRR29 pKa = 11.84RR30 pKa = 11.84NMPALANTRR39 pKa = 11.84IDD41 pKa = 3.38VAGTINALYY50 pKa = 10.23HH51 pKa = 5.24STNISRR57 pKa = 11.84GPDD60 pKa = 3.06SRR62 pKa = 11.84RR63 pKa = 11.84NGPLNSVLSTLNSKK77 pKa = 10.02IPAFFLIRR85 pKa = 11.84TSTFHH90 pKa = 7.08LAADD94 pKa = 3.73HH95 pKa = 6.39FLHH98 pKa = 7.35FEE100 pKa = 4.17GQTATTATATPSPFASILFTISHH123 pKa = 7.07
MM1 pKa = 7.49RR2 pKa = 11.84VDD4 pKa = 3.56IPEE7 pKa = 3.83MGYY10 pKa = 10.35NLVSRR15 pKa = 11.84DD16 pKa = 2.93VDD18 pKa = 5.29LIILLYY24 pKa = 10.58ARR26 pKa = 11.84IWRR29 pKa = 11.84RR30 pKa = 11.84NMPALANTRR39 pKa = 11.84IDD41 pKa = 3.38VAGTINALYY50 pKa = 10.23HH51 pKa = 5.24STNISRR57 pKa = 11.84GPDD60 pKa = 3.06SRR62 pKa = 11.84RR63 pKa = 11.84NGPLNSVLSTLNSKK77 pKa = 10.02IPAFFLIRR85 pKa = 11.84TSTFHH90 pKa = 7.08LAADD94 pKa = 3.73HH95 pKa = 6.39FLHH98 pKa = 7.35FEE100 pKa = 4.17GQTATTATATPSPFASILFTISHH123 pKa = 7.07
Molecular weight: 13.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
837 |
100 |
292 |
167.4 |
19.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.766 ± 0.879 | 1.553 ± 0.619 |
5.018 ± 0.609 | 5.496 ± 1.21 |
3.226 ± 0.557 | 6.571 ± 1.045 |
2.987 ± 0.455 | 6.332 ± 1.126 |
6.93 ± 2.48 | 8.005 ± 1.636 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.389 ± 0.554 | 3.943 ± 0.674 |
3.704 ± 0.679 | 3.226 ± 0.575 |
8.363 ± 0.582 | 6.452 ± 1.414 |
6.332 ± 0.691 | 5.018 ± 0.584 |
1.912 ± 0.507 | 4.421 ± 0.547 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
