
Pteropine orthoreovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Orthoreovirus; unclassified Orthoreovirus
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
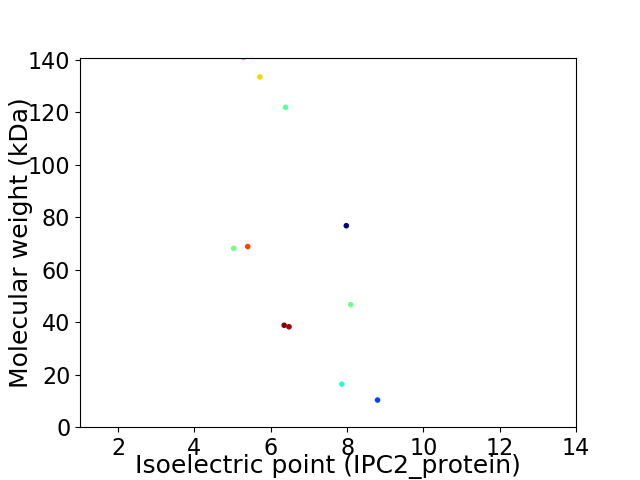
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
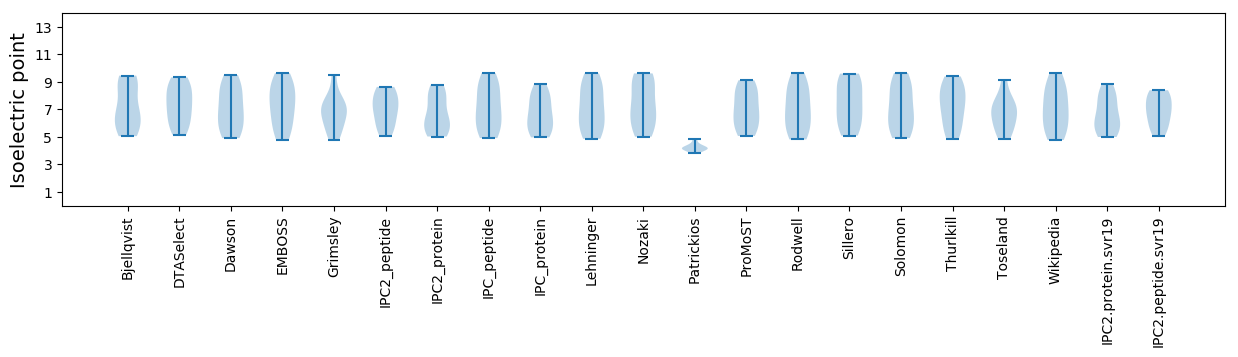
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1CEU6|A0A0A1CEU6_9REOV SNS (Fragment) OS=Pteropine orthoreovirus OX=1459003 PE=4 SV=1
MM1 pKa = 7.32MGNATSIVQNFNIKK15 pKa = 10.61GDD17 pKa = 3.73GNTFSPSAEE26 pKa = 4.24TASSAVPSLSLNPGLLNPGGKK47 pKa = 9.45AWTLIDD53 pKa = 3.53PTVPATAPGSLRR65 pKa = 11.84LMTTEE70 pKa = 4.89DD71 pKa = 3.38VPAALGTSILGSNGAAPSSGMYY93 pKa = 9.91LVPVKK98 pKa = 9.69EE99 pKa = 4.27TLNVVTDD106 pKa = 3.4HH107 pKa = 7.62AIAQFEE113 pKa = 4.42KK114 pKa = 10.71LQSAYY119 pKa = 10.22EE120 pKa = 4.0LDD122 pKa = 3.94RR123 pKa = 11.84DD124 pKa = 3.96YY125 pKa = 11.81LDD127 pKa = 3.99EE128 pKa = 6.08KK129 pKa = 11.05GVAPEE134 pKa = 4.02SVSFKK139 pKa = 11.22NYY141 pKa = 8.17LTYY144 pKa = 10.37VDD146 pKa = 4.96CYY148 pKa = 10.04VGVSARR154 pKa = 11.84QAATNFQKK162 pKa = 10.59HH163 pKa = 5.05VPVVTKK169 pKa = 10.61SKK171 pKa = 9.88MMSFMTSAQNILSLLGPWEE190 pKa = 3.87KK191 pKa = 10.6SIRR194 pKa = 11.84EE195 pKa = 4.29VLTMLPSSTPYY206 pKa = 9.59GTLKK210 pKa = 10.68CDD212 pKa = 3.28MKK214 pKa = 11.18AIVAMLEE221 pKa = 4.01EE222 pKa = 4.3QLPEE226 pKa = 4.29GNLCRR231 pKa = 11.84LYY233 pKa = 10.07PQAAACSIARR243 pKa = 11.84RR244 pKa = 11.84NGGVRR249 pKa = 11.84WKK251 pKa = 10.49EE252 pKa = 4.0PNSDD256 pKa = 3.92EE257 pKa = 4.79APSLATNDD265 pKa = 3.28VAASTMGALANTTPLADD282 pKa = 3.49KK283 pKa = 11.24SNTTEE288 pKa = 3.28EE289 pKa = 4.19SMRR292 pKa = 11.84LVSEE296 pKa = 4.25SSVDD300 pKa = 4.33LICSRR305 pKa = 11.84RR306 pKa = 11.84PISASVWARR315 pKa = 11.84TVEE318 pKa = 3.96PKK320 pKa = 10.41SYY322 pKa = 10.8NIRR325 pKa = 11.84TLKK328 pKa = 10.53VSEE331 pKa = 4.31ALWLRR336 pKa = 11.84QSQVSAGFDD345 pKa = 3.12VAYY348 pKa = 10.49NLNDD352 pKa = 3.08STQRR356 pKa = 11.84HH357 pKa = 4.9FWITTGTTVINLEE370 pKa = 4.19QTGSMMFEE378 pKa = 4.1VNIEE382 pKa = 3.87GKK384 pKa = 9.85DD385 pKa = 3.54YY386 pKa = 11.26KK387 pKa = 10.76KK388 pKa = 11.21GSFQPNGATLVLLVMQSRR406 pKa = 11.84LPFEE410 pKa = 4.04TWTVSSQIEE419 pKa = 4.7GIAQVASIVVTAGSGSTVNTSIIGSTSLSYY449 pKa = 10.94LFEE452 pKa = 6.11RR453 pKa = 11.84EE454 pKa = 4.28TITTANTEE462 pKa = 4.05VNTYY466 pKa = 9.95LLCTWQSSSVCTDD479 pKa = 3.0ADD481 pKa = 3.72SWPDD485 pKa = 2.79AWDD488 pKa = 3.34AVTTLTPLTTGTVTVKK504 pKa = 10.51GSTVEE509 pKa = 3.93KK510 pKa = 9.36VTPVDD515 pKa = 4.81LIGSYY520 pKa = 9.24TPEE523 pKa = 4.28SLTAALPNDD532 pKa = 4.02AGNILALRR540 pKa = 11.84AEE542 pKa = 4.42TLAKK546 pKa = 10.1AVKK549 pKa = 10.6NEE551 pKa = 4.09DD552 pKa = 3.7DD553 pKa = 3.9SVTDD557 pKa = 3.61EE558 pKa = 4.72SSPFSAPIQGILALQQKK575 pKa = 6.91EE576 pKa = 4.66TEE578 pKa = 4.47GTAGTPDD585 pKa = 4.04PKK587 pKa = 10.88PPGTLQKK594 pKa = 9.56IASRR598 pKa = 11.84AMHH601 pKa = 6.47MFLGDD606 pKa = 3.61PKK608 pKa = 10.98SVLKK612 pKa = 9.21QTTPVLKK619 pKa = 10.44DD620 pKa = 3.36PQVWTSFVQGVRR632 pKa = 11.84DD633 pKa = 4.76GIRR636 pKa = 2.98
MM1 pKa = 7.32MGNATSIVQNFNIKK15 pKa = 10.61GDD17 pKa = 3.73GNTFSPSAEE26 pKa = 4.24TASSAVPSLSLNPGLLNPGGKK47 pKa = 9.45AWTLIDD53 pKa = 3.53PTVPATAPGSLRR65 pKa = 11.84LMTTEE70 pKa = 4.89DD71 pKa = 3.38VPAALGTSILGSNGAAPSSGMYY93 pKa = 9.91LVPVKK98 pKa = 9.69EE99 pKa = 4.27TLNVVTDD106 pKa = 3.4HH107 pKa = 7.62AIAQFEE113 pKa = 4.42KK114 pKa = 10.71LQSAYY119 pKa = 10.22EE120 pKa = 4.0LDD122 pKa = 3.94RR123 pKa = 11.84DD124 pKa = 3.96YY125 pKa = 11.81LDD127 pKa = 3.99EE128 pKa = 6.08KK129 pKa = 11.05GVAPEE134 pKa = 4.02SVSFKK139 pKa = 11.22NYY141 pKa = 8.17LTYY144 pKa = 10.37VDD146 pKa = 4.96CYY148 pKa = 10.04VGVSARR154 pKa = 11.84QAATNFQKK162 pKa = 10.59HH163 pKa = 5.05VPVVTKK169 pKa = 10.61SKK171 pKa = 9.88MMSFMTSAQNILSLLGPWEE190 pKa = 3.87KK191 pKa = 10.6SIRR194 pKa = 11.84EE195 pKa = 4.29VLTMLPSSTPYY206 pKa = 9.59GTLKK210 pKa = 10.68CDD212 pKa = 3.28MKK214 pKa = 11.18AIVAMLEE221 pKa = 4.01EE222 pKa = 4.3QLPEE226 pKa = 4.29GNLCRR231 pKa = 11.84LYY233 pKa = 10.07PQAAACSIARR243 pKa = 11.84RR244 pKa = 11.84NGGVRR249 pKa = 11.84WKK251 pKa = 10.49EE252 pKa = 4.0PNSDD256 pKa = 3.92EE257 pKa = 4.79APSLATNDD265 pKa = 3.28VAASTMGALANTTPLADD282 pKa = 3.49KK283 pKa = 11.24SNTTEE288 pKa = 3.28EE289 pKa = 4.19SMRR292 pKa = 11.84LVSEE296 pKa = 4.25SSVDD300 pKa = 4.33LICSRR305 pKa = 11.84RR306 pKa = 11.84PISASVWARR315 pKa = 11.84TVEE318 pKa = 3.96PKK320 pKa = 10.41SYY322 pKa = 10.8NIRR325 pKa = 11.84TLKK328 pKa = 10.53VSEE331 pKa = 4.31ALWLRR336 pKa = 11.84QSQVSAGFDD345 pKa = 3.12VAYY348 pKa = 10.49NLNDD352 pKa = 3.08STQRR356 pKa = 11.84HH357 pKa = 4.9FWITTGTTVINLEE370 pKa = 4.19QTGSMMFEE378 pKa = 4.1VNIEE382 pKa = 3.87GKK384 pKa = 9.85DD385 pKa = 3.54YY386 pKa = 11.26KK387 pKa = 10.76KK388 pKa = 11.21GSFQPNGATLVLLVMQSRR406 pKa = 11.84LPFEE410 pKa = 4.04TWTVSSQIEE419 pKa = 4.7GIAQVASIVVTAGSGSTVNTSIIGSTSLSYY449 pKa = 10.94LFEE452 pKa = 6.11RR453 pKa = 11.84EE454 pKa = 4.28TITTANTEE462 pKa = 4.05VNTYY466 pKa = 9.95LLCTWQSSSVCTDD479 pKa = 3.0ADD481 pKa = 3.72SWPDD485 pKa = 2.79AWDD488 pKa = 3.34AVTTLTPLTTGTVTVKK504 pKa = 10.51GSTVEE509 pKa = 3.93KK510 pKa = 9.36VTPVDD515 pKa = 4.81LIGSYY520 pKa = 9.24TPEE523 pKa = 4.28SLTAALPNDD532 pKa = 4.02AGNILALRR540 pKa = 11.84AEE542 pKa = 4.42TLAKK546 pKa = 10.1AVKK549 pKa = 10.6NEE551 pKa = 4.09DD552 pKa = 3.7DD553 pKa = 3.9SVTDD557 pKa = 3.61EE558 pKa = 4.72SSPFSAPIQGILALQQKK575 pKa = 6.91EE576 pKa = 4.66TEE578 pKa = 4.47GTAGTPDD585 pKa = 4.04PKK587 pKa = 10.88PPGTLQKK594 pKa = 9.56IASRR598 pKa = 11.84AMHH601 pKa = 6.47MFLGDD606 pKa = 3.61PKK608 pKa = 10.98SVLKK612 pKa = 9.21QTTPVLKK619 pKa = 10.44DD620 pKa = 3.36PQVWTSFVQGVRR632 pKa = 11.84DD633 pKa = 4.76GIRR636 pKa = 2.98
Molecular weight: 68.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1CFP6|A0A0A1CFP6_9REOV 1A (Fragment) OS=Pteropine orthoreovirus OX=1459003 PE=4 SV=1
MM1 pKa = 7.66SGDD4 pKa = 3.58CAKK7 pKa = 10.14IVSVFGSVHH16 pKa = 5.7CQSAKK21 pKa = 10.56NSAGGDD27 pKa = 3.61LQATSVFTTYY37 pKa = 9.34WPHH40 pKa = 5.75FAIGGGIIVIILLLGLFYY58 pKa = 10.93CCYY61 pKa = 10.28LKK63 pKa = 10.55WKK65 pKa = 8.3TSQVKK70 pKa = 8.08RR71 pKa = 11.84TYY73 pKa = 10.13RR74 pKa = 11.84RR75 pKa = 11.84EE76 pKa = 4.04LIALTRR82 pKa = 11.84SHH84 pKa = 6.04VHH86 pKa = 6.23PASSGISHH94 pKa = 6.41VV95 pKa = 3.59
MM1 pKa = 7.66SGDD4 pKa = 3.58CAKK7 pKa = 10.14IVSVFGSVHH16 pKa = 5.7CQSAKK21 pKa = 10.56NSAGGDD27 pKa = 3.61LQATSVFTTYY37 pKa = 9.34WPHH40 pKa = 5.75FAIGGGIIVIILLLGLFYY58 pKa = 10.93CCYY61 pKa = 10.28LKK63 pKa = 10.55WKK65 pKa = 8.3TSQVKK70 pKa = 8.08RR71 pKa = 11.84TYY73 pKa = 10.13RR74 pKa = 11.84RR75 pKa = 11.84EE76 pKa = 4.04LIALTRR82 pKa = 11.84SHH84 pKa = 6.04VHH86 pKa = 6.23PASSGISHH94 pKa = 6.41VV95 pKa = 3.59
Molecular weight: 10.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6882 |
95 |
1277 |
625.6 |
69.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.123 ± 0.508 | 1.584 ± 0.124 |
5.812 ± 0.259 | 3.502 ± 0.452 |
4.054 ± 0.294 | 4.693 ± 0.285 |
2.005 ± 0.303 | 4.577 ± 0.258 |
3.124 ± 0.213 | 10.273 ± 0.38 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.318 | 3.909 ± 0.471 |
6.452 ± 0.312 | 3.851 ± 0.278 |
5.696 ± 0.451 | 10.506 ± 0.729 |
6.728 ± 0.436 | 7.774 ± 0.32 |
1.744 ± 0.121 | 2.848 ± 0.338 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
