
Acaromyces ingoldii
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Ustilaginomycotina; Exobasidiomycetes; Exobasidiales; Cryptobasidiaceae; Acaromyces
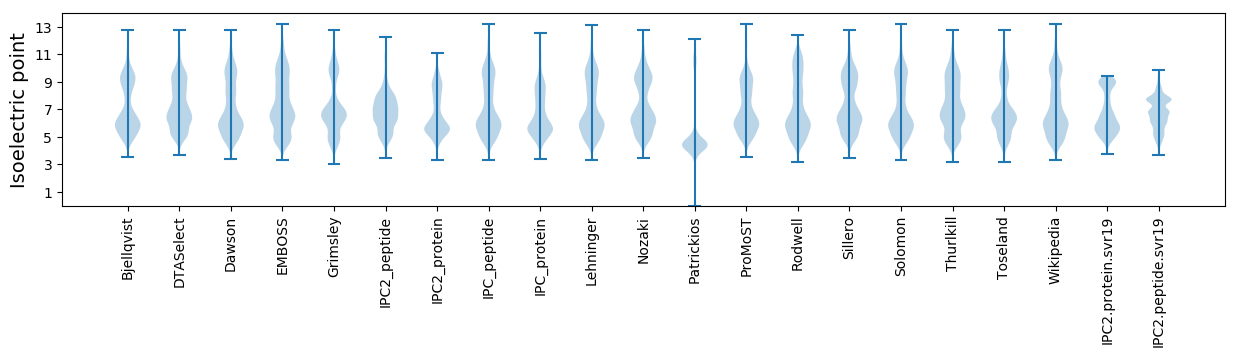
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
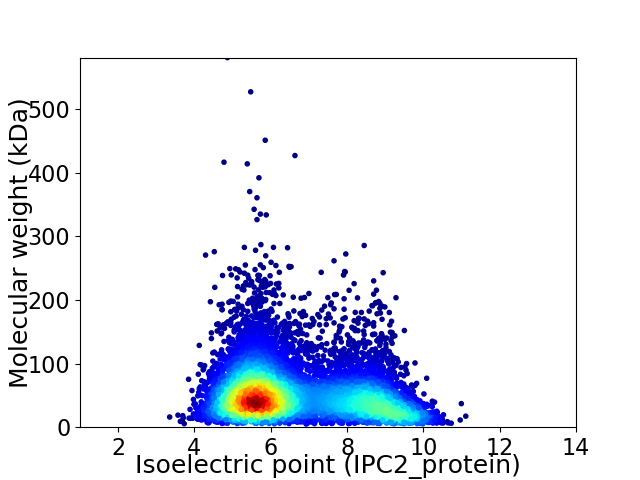
Virtual 2D-PAGE plot for 8019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316YF16|A0A316YF16_9BASI Cir_N domain-containing protein OS=Acaromyces ingoldii OX=215250 GN=FA10DRAFT_174389 PE=3 SV=1
MM1 pKa = 8.02DD2 pKa = 4.47NMINKK7 pKa = 9.47AKK9 pKa = 10.18EE10 pKa = 3.96FASSDD15 pKa = 3.33QGRR18 pKa = 11.84DD19 pKa = 3.28MLNKK23 pKa = 10.35FGGGGNNNNNDD34 pKa = 3.73NNNSGSGGFGSGGNSGSDD52 pKa = 3.37SYY54 pKa = 12.15GSGGNSGNDD63 pKa = 3.33SYY65 pKa = 12.18GSGGNSGNDD74 pKa = 3.33SYY76 pKa = 12.18GSGGNSGSGGGFGGSGGRR94 pKa = 11.84NNDD97 pKa = 3.26DD98 pKa = 3.73SYY100 pKa = 12.15GGGNSGSGGGFGGNDD115 pKa = 3.36STSSGGFGGNNQGDD129 pKa = 4.47SYY131 pKa = 11.75SSGNQRR137 pKa = 11.84GGSGGGQFGSSDD149 pKa = 3.42SDD151 pKa = 3.89SYY153 pKa = 12.12GSGNQQGGGGGFGSGNQGGNDD174 pKa = 3.48SYY176 pKa = 12.08GSGNQGSGGYY186 pKa = 10.3GGGNQDD192 pKa = 3.47NYY194 pKa = 11.75
MM1 pKa = 8.02DD2 pKa = 4.47NMINKK7 pKa = 9.47AKK9 pKa = 10.18EE10 pKa = 3.96FASSDD15 pKa = 3.33QGRR18 pKa = 11.84DD19 pKa = 3.28MLNKK23 pKa = 10.35FGGGGNNNNNDD34 pKa = 3.73NNNSGSGGFGSGGNSGSDD52 pKa = 3.37SYY54 pKa = 12.15GSGGNSGNDD63 pKa = 3.33SYY65 pKa = 12.18GSGGNSGNDD74 pKa = 3.33SYY76 pKa = 12.18GSGGNSGSGGGFGGSGGRR94 pKa = 11.84NNDD97 pKa = 3.26DD98 pKa = 3.73SYY100 pKa = 12.15GGGNSGSGGGFGGNDD115 pKa = 3.36STSSGGFGGNNQGDD129 pKa = 4.47SYY131 pKa = 11.75SSGNQRR137 pKa = 11.84GGSGGGQFGSSDD149 pKa = 3.42SDD151 pKa = 3.89SYY153 pKa = 12.12GSGNQQGGGGGFGSGNQGGNDD174 pKa = 3.48SYY176 pKa = 12.08GSGNQGSGGYY186 pKa = 10.3GGGNQDD192 pKa = 3.47NYY194 pKa = 11.75
Molecular weight: 18.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316YWW8|A0A316YWW8_9BASI Histone H3 OS=Acaromyces ingoldii OX=215250 GN=FA10DRAFT_237339 PE=3 SV=1
MM1 pKa = 8.11DD2 pKa = 4.94SALQEE7 pKa = 4.12QIRR10 pKa = 11.84KK11 pKa = 9.69GKK13 pKa = 9.37GLKK16 pKa = 8.44KK17 pKa = 8.67TQTNDD22 pKa = 2.77RR23 pKa = 11.84SKK25 pKa = 10.5PAVAGAVGGSGGGGGGSGGSGLAKK49 pKa = 10.33APPVPGGGGAAAAPSSGGPPALAGLFAGGMPKK81 pKa = 10.45LKK83 pKa = 10.39PSGGPGAPPPTVPAARR99 pKa = 11.84PPRR102 pKa = 11.84PPGSAAPPPPAPPAPAAPKK121 pKa = 10.13RR122 pKa = 11.84PGGAPPPPPPPPAPASSVATPPRR145 pKa = 11.84TAPPPPRR152 pKa = 11.84PPGRR156 pKa = 11.84GPPAPTSAPPPPRR169 pKa = 11.84PPPSLPGRR177 pKa = 11.84APAAPPAPPPPPPPASSSPAPPARR201 pKa = 11.84TVPQAPAPPKK211 pKa = 10.16RR212 pKa = 11.84PGGAAPPPPPSRR224 pKa = 11.84PMSSSSTVGRR234 pKa = 11.84TVPPPPPGRR243 pKa = 11.84GSPSSPSTAGPTRR256 pKa = 11.84SVPPPPPRR264 pKa = 11.84ASAASSAPPPAPPRR278 pKa = 11.84APPAPSRR285 pKa = 11.84MPAAPPPPAPQRR297 pKa = 11.84GPAAPPPTAPSRR309 pKa = 11.84APAAPPPPAPSRR321 pKa = 11.84APATPPSRR329 pKa = 11.84TPAAPPPPPPGRR341 pKa = 11.84APPPTAAAPPPPSAAPPAPSSRR363 pKa = 11.84ASLQPVARR371 pKa = 11.84ANGFGTVGNGTGNGTASSTAPSGKK395 pKa = 10.05SVV397 pKa = 2.87
MM1 pKa = 8.11DD2 pKa = 4.94SALQEE7 pKa = 4.12QIRR10 pKa = 11.84KK11 pKa = 9.69GKK13 pKa = 9.37GLKK16 pKa = 8.44KK17 pKa = 8.67TQTNDD22 pKa = 2.77RR23 pKa = 11.84SKK25 pKa = 10.5PAVAGAVGGSGGGGGGSGGSGLAKK49 pKa = 10.33APPVPGGGGAAAAPSSGGPPALAGLFAGGMPKK81 pKa = 10.45LKK83 pKa = 10.39PSGGPGAPPPTVPAARR99 pKa = 11.84PPRR102 pKa = 11.84PPGSAAPPPPAPPAPAAPKK121 pKa = 10.13RR122 pKa = 11.84PGGAPPPPPPPPAPASSVATPPRR145 pKa = 11.84TAPPPPRR152 pKa = 11.84PPGRR156 pKa = 11.84GPPAPTSAPPPPRR169 pKa = 11.84PPPSLPGRR177 pKa = 11.84APAAPPAPPPPPPPASSSPAPPARR201 pKa = 11.84TVPQAPAPPKK211 pKa = 10.16RR212 pKa = 11.84PGGAAPPPPPSRR224 pKa = 11.84PMSSSSTVGRR234 pKa = 11.84TVPPPPPGRR243 pKa = 11.84GSPSSPSTAGPTRR256 pKa = 11.84SVPPPPPRR264 pKa = 11.84ASAASSAPPPAPPRR278 pKa = 11.84APPAPSRR285 pKa = 11.84MPAAPPPPAPQRR297 pKa = 11.84GPAAPPPTAPSRR309 pKa = 11.84APAAPPPPAPSRR321 pKa = 11.84APATPPSRR329 pKa = 11.84TPAAPPPPPPGRR341 pKa = 11.84APPPTAAAPPPPSAAPPAPSSRR363 pKa = 11.84ASLQPVARR371 pKa = 11.84ANGFGTVGNGTGNGTASSTAPSGKK395 pKa = 10.05SVV397 pKa = 2.87
Molecular weight: 37.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4210363 |
49 |
5290 |
525.0 |
57.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.379 ± 0.03 | 1.049 ± 0.009 |
5.777 ± 0.019 | 6.219 ± 0.026 |
3.252 ± 0.016 | 7.648 ± 0.025 |
2.38 ± 0.012 | 3.777 ± 0.018 |
4.515 ± 0.025 | 8.832 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.993 ± 0.008 | 2.933 ± 0.014 |
5.849 ± 0.028 | 4.328 ± 0.025 |
6.784 ± 0.024 | 9.301 ± 0.044 |
5.536 ± 0.019 | 6.0 ± 0.022 |
1.263 ± 0.009 | 2.186 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
