
Motiliproteus coralliicola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Motiliproteus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
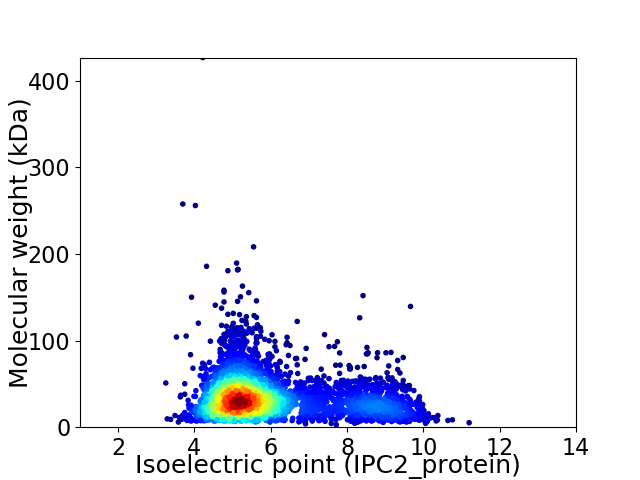
Virtual 2D-PAGE plot for 3762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369WLB1|A0A369WLB1_9GAMM NCS2 family permease OS=Motiliproteus coralliicola OX=2283196 GN=DV711_09930 PE=3 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84VINTGRR8 pKa = 11.84SNRR11 pKa = 11.84LLTVILQLGLLMLLAASAQAGRR33 pKa = 11.84TINSATVNGGSSATVAPGASITVTLNVTTDD63 pKa = 3.61GNGSNNDD70 pKa = 3.4WEE72 pKa = 4.59STDD75 pKa = 3.24WLIGTSSSGSYY86 pKa = 8.5TCNNHH91 pKa = 6.75ADD93 pKa = 3.48NTFTGTHH100 pKa = 5.68TEE102 pKa = 4.1SFTITAPSTPGTYY115 pKa = 9.82NAYY118 pKa = 9.93FWASHH123 pKa = 6.17NDD125 pKa = 3.1SCGGSGSNLFTLTNAITVSTGTTCSAEE152 pKa = 4.17TVADD156 pKa = 3.76NFDD159 pKa = 3.78SVSFSQNDD167 pKa = 3.7GSRR170 pKa = 11.84NWSGDD175 pKa = 2.74WQEE178 pKa = 4.35IGEE181 pKa = 4.41SDD183 pKa = 3.7GASAGFVRR191 pKa = 11.84VRR193 pKa = 11.84NDD195 pKa = 2.69SCSSGNCLRR204 pKa = 11.84LGVISSEE211 pKa = 4.07PAQSFSNVGASRR223 pKa = 11.84EE224 pKa = 3.93VDD226 pKa = 3.28LSGATTATLTFNYY239 pKa = 7.74RR240 pKa = 11.84TGVIQNSEE248 pKa = 4.4TVSLQVSANGGSSWTTLRR266 pKa = 11.84TYY268 pKa = 9.23TVSSTNSSATAEE280 pKa = 4.29SVDD283 pKa = 3.72LSSYY287 pKa = 9.92IASNTQIRR295 pKa = 11.84FLASGSNAIVGFYY308 pKa = 10.71ADD310 pKa = 5.54DD311 pKa = 3.78IEE313 pKa = 5.3ISYY316 pKa = 10.28DD317 pKa = 3.41PCSTLLAEE325 pKa = 4.27WRR327 pKa = 11.84LDD329 pKa = 3.43EE330 pKa = 4.35TSWNGTSDD338 pKa = 3.54EE339 pKa = 4.45VVDD342 pKa = 5.03SEE344 pKa = 4.63GSNHH348 pKa = 5.65GVANSADD355 pKa = 3.78TASGLLCNAADD366 pKa = 4.53LSAAGTSDD374 pKa = 4.09YY375 pKa = 11.17ISLGSGALNGATDD388 pKa = 3.97FTISAWIKK396 pKa = 8.44TSNSGSQALISGANSSQANEE416 pKa = 4.3LIMWFPSATSFEE428 pKa = 4.28PWLKK432 pKa = 10.78GVDD435 pKa = 3.7PSSISIGNIADD446 pKa = 4.07NQWHH450 pKa = 6.23HH451 pKa = 6.49LVWTRR456 pKa = 11.84SGSQNCIYY464 pKa = 10.44RR465 pKa = 11.84DD466 pKa = 3.62ASLQGCTSGSTSALNIDD483 pKa = 3.72SGGLILGQEE492 pKa = 4.09QDD494 pKa = 3.65SVGGGFDD501 pKa = 5.36LGQDD505 pKa = 2.8WRR507 pKa = 11.84GLVDD511 pKa = 3.9EE512 pKa = 5.24LLIFNEE518 pKa = 4.53AVSASQITSIYY529 pKa = 10.26NNTLAGDD536 pKa = 3.53NWDD539 pKa = 3.56GTEE542 pKa = 4.0RR543 pKa = 11.84DD544 pKa = 4.59CPSGSCSVTYY554 pKa = 10.94SDD556 pKa = 5.35DD557 pKa = 3.32FSAASFTNSSGSVNWSGGWTEE578 pKa = 5.21HH579 pKa = 6.61DD580 pKa = 3.7TASTGAAAGYY590 pKa = 10.25VSISGGKK597 pKa = 8.97LHH599 pKa = 6.55LQNYY603 pKa = 6.65TPSEE607 pKa = 4.16SSSRR611 pKa = 11.84PGVARR616 pKa = 11.84GFSLADD622 pKa = 3.25ASEE625 pKa = 4.39AVVSYY630 pKa = 10.63SYY632 pKa = 11.63SLTDD636 pKa = 4.79AVDD639 pKa = 3.54SDD641 pKa = 4.47DD642 pKa = 3.95SVLVEE647 pKa = 4.24ISDD650 pKa = 4.61DD651 pKa = 4.28GGSSWTTIDD660 pKa = 3.93TITGLGGGSHH670 pKa = 6.68SNSISLSSFSSISFTNNMQISIRR693 pKa = 11.84ISGNINSGSCCYY705 pKa = 10.47GGSNEE710 pKa = 4.19IISFEE715 pKa = 4.14FVTITTQEE723 pKa = 3.98PCSASDD729 pKa = 3.32VDD731 pKa = 4.92HH732 pKa = 6.43YY733 pKa = 11.42AISHH737 pKa = 6.18SGQGVTCEE745 pKa = 4.56AEE747 pKa = 4.46SITITAHH754 pKa = 6.62DD755 pKa = 3.98SSEE758 pKa = 4.12GTVEE762 pKa = 4.11PGNSVSLTLNTGTGIGDD779 pKa = 3.37WSLITGNGSLANGTAGDD796 pKa = 5.06GIASYY801 pKa = 10.63SWGSGEE807 pKa = 4.24SAVVLALAHH816 pKa = 6.28PAVTTAPHH824 pKa = 6.14MDD826 pKa = 3.27IDD828 pKa = 4.77LVDD831 pKa = 4.05NNSATDD837 pKa = 3.53KK838 pKa = 11.41DD839 pKa = 3.96GDD841 pKa = 3.87GTEE844 pKa = 4.11DD845 pKa = 3.41PKK847 pKa = 11.32LAFSDD852 pKa = 3.31AGFRR856 pKa = 11.84FYY858 pKa = 11.39GAGVANNIGNQIAGKK873 pKa = 9.54EE874 pKa = 4.26SNLAPGNQTIEE885 pKa = 3.71IRR887 pKa = 11.84AVQTNTDD894 pKa = 3.22TGACEE899 pKa = 4.04ARR901 pKa = 11.84LTNTQIVQMAYY912 pKa = 8.73QCRR915 pKa = 11.84NPATCQGDD923 pKa = 3.56NKK925 pKa = 9.94LTLNDD930 pKa = 3.63STTLRR935 pKa = 11.84ANPATGVTNYY945 pKa = 10.9SNVSLNFGSNGQASFTFDD963 pKa = 3.27YY964 pKa = 11.04SDD966 pKa = 3.72AGLIQLHH973 pKa = 6.21ASKK976 pKa = 9.33TLAASGEE983 pKa = 4.26DD984 pKa = 3.6PQVTLSGSSNEE995 pKa = 3.9FVVRR999 pKa = 11.84PFGFHH1004 pKa = 6.54IDD1006 pKa = 3.71VPGNPAAISAAGDD1019 pKa = 3.55PFKK1022 pKa = 11.05AAGEE1026 pKa = 4.09AFAATVTAVLWASGDD1041 pKa = 3.84DD1042 pKa = 3.81SDD1044 pKa = 5.94SDD1046 pKa = 4.14GVPDD1050 pKa = 4.68SGADD1054 pKa = 3.42LSDD1057 pKa = 3.63NGVTANFGQEE1067 pKa = 3.94STAEE1071 pKa = 4.36TVTLTQGLVLPTNGTNPALSNGTFSGFSAGASTKK1105 pKa = 10.96SNLSWSEE1112 pKa = 3.61VGIISLTANVTDD1124 pKa = 4.65GSYY1127 pKa = 10.75LGAGAVTGTLPYY1139 pKa = 10.15VGRR1142 pKa = 11.84FYY1144 pKa = 10.59PSHH1147 pKa = 6.48FRR1149 pKa = 11.84LSAATLTNRR1158 pKa = 11.84TDD1160 pKa = 3.54LASCSSSSFTYY1171 pKa = 9.04MGEE1174 pKa = 4.06PFGVQYY1180 pKa = 10.96SLSAEE1185 pKa = 4.06NSSGVITRR1193 pKa = 11.84NYY1195 pKa = 9.89AGNFAKK1201 pKa = 10.6LDD1203 pKa = 3.62TSGEE1207 pKa = 4.21LNLGAVDD1214 pKa = 4.01GTTDD1218 pKa = 3.5LSSRR1222 pKa = 11.84LTLDD1226 pKa = 2.92SASIVWPALDD1236 pKa = 3.4HH1237 pKa = 6.46TGAGTAAIDD1246 pKa = 3.5LNLRR1250 pKa = 11.84IDD1252 pKa = 4.09RR1253 pKa = 11.84DD1254 pKa = 4.06SLPDD1258 pKa = 3.58GPFAALSVGTAPNDD1272 pKa = 3.04EE1273 pKa = 4.26DD1274 pKa = 4.0AQLNSFDD1281 pKa = 5.49LDD1283 pKa = 3.7VSGDD1287 pKa = 3.58TFFDD1291 pKa = 3.53HH1292 pKa = 7.11GLLGSTRR1299 pKa = 11.84IRR1301 pKa = 11.84YY1302 pKa = 8.32GRR1304 pKa = 11.84LLVGNTFGPEE1314 pKa = 4.01TEE1316 pKa = 4.34DD1317 pKa = 5.02LEE1319 pKa = 5.13VPLQLEE1325 pKa = 4.44YY1326 pKa = 10.87FDD1328 pKa = 5.4GINFILNTDD1337 pKa = 4.26DD1338 pKa = 4.9SCTTYY1343 pKa = 10.98INTSTSLDD1351 pKa = 3.37HH1352 pKa = 7.34SNYY1355 pKa = 10.38GSDD1358 pKa = 3.31NLEE1361 pKa = 4.4AGDD1364 pKa = 4.16TSVTSPNTEE1373 pKa = 4.01TEE1375 pKa = 4.48VVAGQSPSSAPLVLSAPGVGNEE1397 pKa = 4.12GQVDD1401 pKa = 5.52LIYY1404 pKa = 10.6AAPSWLEE1411 pKa = 3.58FDD1413 pKa = 3.53WFGGGDD1419 pKa = 3.58TDD1421 pKa = 4.84PRR1423 pKa = 11.84GQATFGRR1430 pKa = 11.84YY1431 pKa = 9.06RR1432 pKa = 11.84GNDD1435 pKa = 2.79RR1436 pKa = 11.84MIFWQEE1442 pKa = 3.56NFSQQ1446 pKa = 3.6
MM1 pKa = 7.55RR2 pKa = 11.84VINTGRR8 pKa = 11.84SNRR11 pKa = 11.84LLTVILQLGLLMLLAASAQAGRR33 pKa = 11.84TINSATVNGGSSATVAPGASITVTLNVTTDD63 pKa = 3.61GNGSNNDD70 pKa = 3.4WEE72 pKa = 4.59STDD75 pKa = 3.24WLIGTSSSGSYY86 pKa = 8.5TCNNHH91 pKa = 6.75ADD93 pKa = 3.48NTFTGTHH100 pKa = 5.68TEE102 pKa = 4.1SFTITAPSTPGTYY115 pKa = 9.82NAYY118 pKa = 9.93FWASHH123 pKa = 6.17NDD125 pKa = 3.1SCGGSGSNLFTLTNAITVSTGTTCSAEE152 pKa = 4.17TVADD156 pKa = 3.76NFDD159 pKa = 3.78SVSFSQNDD167 pKa = 3.7GSRR170 pKa = 11.84NWSGDD175 pKa = 2.74WQEE178 pKa = 4.35IGEE181 pKa = 4.41SDD183 pKa = 3.7GASAGFVRR191 pKa = 11.84VRR193 pKa = 11.84NDD195 pKa = 2.69SCSSGNCLRR204 pKa = 11.84LGVISSEE211 pKa = 4.07PAQSFSNVGASRR223 pKa = 11.84EE224 pKa = 3.93VDD226 pKa = 3.28LSGATTATLTFNYY239 pKa = 7.74RR240 pKa = 11.84TGVIQNSEE248 pKa = 4.4TVSLQVSANGGSSWTTLRR266 pKa = 11.84TYY268 pKa = 9.23TVSSTNSSATAEE280 pKa = 4.29SVDD283 pKa = 3.72LSSYY287 pKa = 9.92IASNTQIRR295 pKa = 11.84FLASGSNAIVGFYY308 pKa = 10.71ADD310 pKa = 5.54DD311 pKa = 3.78IEE313 pKa = 5.3ISYY316 pKa = 10.28DD317 pKa = 3.41PCSTLLAEE325 pKa = 4.27WRR327 pKa = 11.84LDD329 pKa = 3.43EE330 pKa = 4.35TSWNGTSDD338 pKa = 3.54EE339 pKa = 4.45VVDD342 pKa = 5.03SEE344 pKa = 4.63GSNHH348 pKa = 5.65GVANSADD355 pKa = 3.78TASGLLCNAADD366 pKa = 4.53LSAAGTSDD374 pKa = 4.09YY375 pKa = 11.17ISLGSGALNGATDD388 pKa = 3.97FTISAWIKK396 pKa = 8.44TSNSGSQALISGANSSQANEE416 pKa = 4.3LIMWFPSATSFEE428 pKa = 4.28PWLKK432 pKa = 10.78GVDD435 pKa = 3.7PSSISIGNIADD446 pKa = 4.07NQWHH450 pKa = 6.23HH451 pKa = 6.49LVWTRR456 pKa = 11.84SGSQNCIYY464 pKa = 10.44RR465 pKa = 11.84DD466 pKa = 3.62ASLQGCTSGSTSALNIDD483 pKa = 3.72SGGLILGQEE492 pKa = 4.09QDD494 pKa = 3.65SVGGGFDD501 pKa = 5.36LGQDD505 pKa = 2.8WRR507 pKa = 11.84GLVDD511 pKa = 3.9EE512 pKa = 5.24LLIFNEE518 pKa = 4.53AVSASQITSIYY529 pKa = 10.26NNTLAGDD536 pKa = 3.53NWDD539 pKa = 3.56GTEE542 pKa = 4.0RR543 pKa = 11.84DD544 pKa = 4.59CPSGSCSVTYY554 pKa = 10.94SDD556 pKa = 5.35DD557 pKa = 3.32FSAASFTNSSGSVNWSGGWTEE578 pKa = 5.21HH579 pKa = 6.61DD580 pKa = 3.7TASTGAAAGYY590 pKa = 10.25VSISGGKK597 pKa = 8.97LHH599 pKa = 6.55LQNYY603 pKa = 6.65TPSEE607 pKa = 4.16SSSRR611 pKa = 11.84PGVARR616 pKa = 11.84GFSLADD622 pKa = 3.25ASEE625 pKa = 4.39AVVSYY630 pKa = 10.63SYY632 pKa = 11.63SLTDD636 pKa = 4.79AVDD639 pKa = 3.54SDD641 pKa = 4.47DD642 pKa = 3.95SVLVEE647 pKa = 4.24ISDD650 pKa = 4.61DD651 pKa = 4.28GGSSWTTIDD660 pKa = 3.93TITGLGGGSHH670 pKa = 6.68SNSISLSSFSSISFTNNMQISIRR693 pKa = 11.84ISGNINSGSCCYY705 pKa = 10.47GGSNEE710 pKa = 4.19IISFEE715 pKa = 4.14FVTITTQEE723 pKa = 3.98PCSASDD729 pKa = 3.32VDD731 pKa = 4.92HH732 pKa = 6.43YY733 pKa = 11.42AISHH737 pKa = 6.18SGQGVTCEE745 pKa = 4.56AEE747 pKa = 4.46SITITAHH754 pKa = 6.62DD755 pKa = 3.98SSEE758 pKa = 4.12GTVEE762 pKa = 4.11PGNSVSLTLNTGTGIGDD779 pKa = 3.37WSLITGNGSLANGTAGDD796 pKa = 5.06GIASYY801 pKa = 10.63SWGSGEE807 pKa = 4.24SAVVLALAHH816 pKa = 6.28PAVTTAPHH824 pKa = 6.14MDD826 pKa = 3.27IDD828 pKa = 4.77LVDD831 pKa = 4.05NNSATDD837 pKa = 3.53KK838 pKa = 11.41DD839 pKa = 3.96GDD841 pKa = 3.87GTEE844 pKa = 4.11DD845 pKa = 3.41PKK847 pKa = 11.32LAFSDD852 pKa = 3.31AGFRR856 pKa = 11.84FYY858 pKa = 11.39GAGVANNIGNQIAGKK873 pKa = 9.54EE874 pKa = 4.26SNLAPGNQTIEE885 pKa = 3.71IRR887 pKa = 11.84AVQTNTDD894 pKa = 3.22TGACEE899 pKa = 4.04ARR901 pKa = 11.84LTNTQIVQMAYY912 pKa = 8.73QCRR915 pKa = 11.84NPATCQGDD923 pKa = 3.56NKK925 pKa = 9.94LTLNDD930 pKa = 3.63STTLRR935 pKa = 11.84ANPATGVTNYY945 pKa = 10.9SNVSLNFGSNGQASFTFDD963 pKa = 3.27YY964 pKa = 11.04SDD966 pKa = 3.72AGLIQLHH973 pKa = 6.21ASKK976 pKa = 9.33TLAASGEE983 pKa = 4.26DD984 pKa = 3.6PQVTLSGSSNEE995 pKa = 3.9FVVRR999 pKa = 11.84PFGFHH1004 pKa = 6.54IDD1006 pKa = 3.71VPGNPAAISAAGDD1019 pKa = 3.55PFKK1022 pKa = 11.05AAGEE1026 pKa = 4.09AFAATVTAVLWASGDD1041 pKa = 3.84DD1042 pKa = 3.81SDD1044 pKa = 5.94SDD1046 pKa = 4.14GVPDD1050 pKa = 4.68SGADD1054 pKa = 3.42LSDD1057 pKa = 3.63NGVTANFGQEE1067 pKa = 3.94STAEE1071 pKa = 4.36TVTLTQGLVLPTNGTNPALSNGTFSGFSAGASTKK1105 pKa = 10.96SNLSWSEE1112 pKa = 3.61VGIISLTANVTDD1124 pKa = 4.65GSYY1127 pKa = 10.75LGAGAVTGTLPYY1139 pKa = 10.15VGRR1142 pKa = 11.84FYY1144 pKa = 10.59PSHH1147 pKa = 6.48FRR1149 pKa = 11.84LSAATLTNRR1158 pKa = 11.84TDD1160 pKa = 3.54LASCSSSSFTYY1171 pKa = 9.04MGEE1174 pKa = 4.06PFGVQYY1180 pKa = 10.96SLSAEE1185 pKa = 4.06NSSGVITRR1193 pKa = 11.84NYY1195 pKa = 9.89AGNFAKK1201 pKa = 10.6LDD1203 pKa = 3.62TSGEE1207 pKa = 4.21LNLGAVDD1214 pKa = 4.01GTTDD1218 pKa = 3.5LSSRR1222 pKa = 11.84LTLDD1226 pKa = 2.92SASIVWPALDD1236 pKa = 3.4HH1237 pKa = 6.46TGAGTAAIDD1246 pKa = 3.5LNLRR1250 pKa = 11.84IDD1252 pKa = 4.09RR1253 pKa = 11.84DD1254 pKa = 4.06SLPDD1258 pKa = 3.58GPFAALSVGTAPNDD1272 pKa = 3.04EE1273 pKa = 4.26DD1274 pKa = 4.0AQLNSFDD1281 pKa = 5.49LDD1283 pKa = 3.7VSGDD1287 pKa = 3.58TFFDD1291 pKa = 3.53HH1292 pKa = 7.11GLLGSTRR1299 pKa = 11.84IRR1301 pKa = 11.84YY1302 pKa = 8.32GRR1304 pKa = 11.84LLVGNTFGPEE1314 pKa = 4.01TEE1316 pKa = 4.34DD1317 pKa = 5.02LEE1319 pKa = 5.13VPLQLEE1325 pKa = 4.44YY1326 pKa = 10.87FDD1328 pKa = 5.4GINFILNTDD1337 pKa = 4.26DD1338 pKa = 4.9SCTTYY1343 pKa = 10.98INTSTSLDD1351 pKa = 3.37HH1352 pKa = 7.34SNYY1355 pKa = 10.38GSDD1358 pKa = 3.31NLEE1361 pKa = 4.4AGDD1364 pKa = 4.16TSVTSPNTEE1373 pKa = 4.01TEE1375 pKa = 4.48VVAGQSPSSAPLVLSAPGVGNEE1397 pKa = 4.12GQVDD1401 pKa = 5.52LIYY1404 pKa = 10.6AAPSWLEE1411 pKa = 3.58FDD1413 pKa = 3.53WFGGGDD1419 pKa = 3.58TDD1421 pKa = 4.84PRR1423 pKa = 11.84GQATFGRR1430 pKa = 11.84YY1431 pKa = 9.06RR1432 pKa = 11.84GNDD1435 pKa = 2.79RR1436 pKa = 11.84MIFWQEE1442 pKa = 3.56NFSQQ1446 pKa = 3.6
Molecular weight: 150.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369WA03|A0A369WA03_9GAMM Cupin domain-containing protein OS=Motiliproteus coralliicola OX=2283196 GN=DV711_14570 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPSVLKK12 pKa = 10.46RR13 pKa = 11.84ARR15 pKa = 11.84NHH17 pKa = 5.26GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.21SGRR29 pKa = 11.84QVLTRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.12GRR40 pKa = 11.84KK41 pKa = 8.87SLSAA45 pKa = 3.86
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPSVLKK12 pKa = 10.46RR13 pKa = 11.84ARR15 pKa = 11.84NHH17 pKa = 5.26GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.21SGRR29 pKa = 11.84QVLTRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.12GRR40 pKa = 11.84KK41 pKa = 8.87SLSAA45 pKa = 3.86
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1227948 |
23 |
3900 |
326.4 |
36.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.433 ± 0.042 | 1.09 ± 0.016 |
5.729 ± 0.035 | 6.335 ± 0.04 |
3.636 ± 0.027 | 7.434 ± 0.034 |
2.191 ± 0.017 | 5.452 ± 0.035 |
3.834 ± 0.033 | 11.507 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.02 | 3.333 ± 0.024 |
4.566 ± 0.027 | 5.443 ± 0.045 |
5.948 ± 0.036 | 6.284 ± 0.031 |
4.701 ± 0.032 | 6.675 ± 0.034 |
1.28 ± 0.015 | 2.692 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
