
Myxococcales bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; unclassified Myxococcales
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
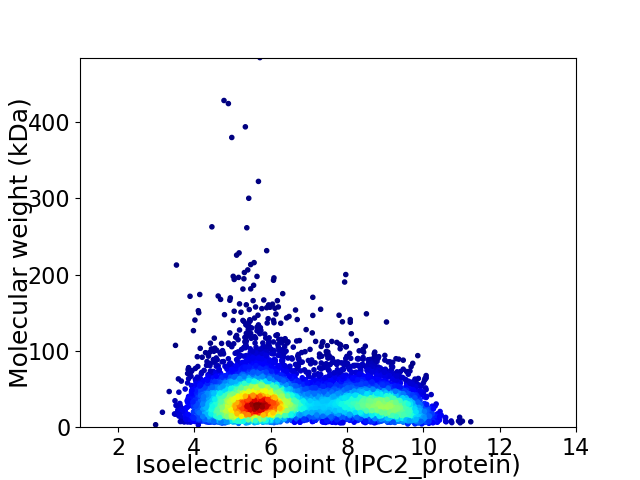
Virtual 2D-PAGE plot for 7151 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q5X5Z9|A0A4Q5X5Z9_9DELT Antibiotic biosynthesis monooxygenase OS=Myxococcales bacterium OX=2026763 GN=EOO73_25305 PE=4 SV=1
MM1 pKa = 7.28TKK3 pKa = 10.22SISAIFLTCAVIATQGTLNAAVRR26 pKa = 11.84SDD28 pKa = 3.09IVALLEE34 pKa = 3.83VDD36 pKa = 3.8PAGSTLVVVDD46 pKa = 5.11GSAVNSTIKK55 pKa = 10.43AGKK58 pKa = 9.86VVLQANDD65 pKa = 4.14PNCDD69 pKa = 3.63PNVAPPCSYY78 pKa = 10.86VLKK81 pKa = 10.54KK82 pKa = 10.62LVIEE86 pKa = 4.26VTDD89 pKa = 4.25FSFKK93 pKa = 11.0DD94 pKa = 3.48EE95 pKa = 4.29NVTNGQFITTGPVNVVDD112 pKa = 4.34DD113 pKa = 4.33GGGIRR118 pKa = 11.84LPAGTPIGFAVTVGDD133 pKa = 4.53DD134 pKa = 3.22RR135 pKa = 11.84AGGIGSVAAPVLIQLDD151 pKa = 3.85VASQQMSLTGSFQGSLEE168 pKa = 4.13GFDD171 pKa = 3.25VAAAVAAQAQSPFVNVPPKK190 pKa = 10.41ISIEE194 pKa = 4.03KK195 pKa = 8.69TLQALEE201 pKa = 4.75CEE203 pKa = 5.06PIPLVASVTDD213 pKa = 3.76ATPAPNDD220 pKa = 3.99DD221 pKa = 3.54VFAPAWAIDD230 pKa = 3.61GVNQGFAPAGFASLAPGAHH249 pKa = 6.09FVQASVQDD257 pKa = 3.67FFGALAVDD265 pKa = 3.84SFVVNVSPDD274 pKa = 3.21EE275 pKa = 4.39APPVFTFVPSDD286 pKa = 3.39IKK288 pKa = 10.27TSSCGGLNIGVATAVDD304 pKa = 3.32NCGEE308 pKa = 4.2VEE310 pKa = 4.41VTSDD314 pKa = 3.81APAKK318 pKa = 9.6FAAGTTVVSWTARR331 pKa = 11.84DD332 pKa = 3.38ARR334 pKa = 11.84GNEE337 pKa = 3.88TVAQQVVTASLKK349 pKa = 10.83DD350 pKa = 3.72DD351 pKa = 4.6ASCCPVGSNVLVGSANNDD369 pKa = 3.32NLVGSSGPDD378 pKa = 3.26CILGLRR384 pKa = 11.84GQDD387 pKa = 3.42QIDD390 pKa = 3.76GGGGDD395 pKa = 4.46DD396 pKa = 4.71VISGGDD402 pKa = 3.26GDD404 pKa = 5.43DD405 pKa = 3.45ILRR408 pKa = 11.84AGAGNDD414 pKa = 4.0LVWAGPGQDD423 pKa = 3.58QLFGGIGDD431 pKa = 4.97DD432 pKa = 4.32LLSGNDD438 pKa = 3.6GDD440 pKa = 5.0DD441 pKa = 3.6LLRR444 pKa = 11.84GEE446 pKa = 5.14GGLDD450 pKa = 3.3TLVGGQGQDD459 pKa = 2.63ILYY462 pKa = 10.75GGDD465 pKa = 4.05DD466 pKa = 3.69NDD468 pKa = 3.97ILTGEE473 pKa = 4.71AGDD476 pKa = 3.95DD477 pKa = 3.62QLFGEE482 pKa = 5.07AGDD485 pKa = 4.49DD486 pKa = 4.0VLDD489 pKa = 5.07GGDD492 pKa = 4.31NNDD495 pKa = 3.32KK496 pKa = 10.01CTGGSGYY503 pKa = 9.05DD504 pKa = 3.38TMLSCTALDD513 pKa = 3.61TLEE516 pKa = 4.79NPGPPSFPGSPSYY529 pKa = 9.94EE530 pKa = 4.06VCEE533 pKa = 4.16CRR535 pKa = 11.84PNKK538 pKa = 10.31CADD541 pKa = 3.47CAAPVATCDD550 pKa = 4.04ASNGCAQIVQCVRR563 pKa = 11.84QAANCNLPHH572 pKa = 6.62EE573 pKa = 4.91CSALCEE579 pKa = 4.28SGRR582 pKa = 11.84TQSAIGAARR591 pKa = 11.84QLASCFGGCC600 pKa = 4.91
MM1 pKa = 7.28TKK3 pKa = 10.22SISAIFLTCAVIATQGTLNAAVRR26 pKa = 11.84SDD28 pKa = 3.09IVALLEE34 pKa = 3.83VDD36 pKa = 3.8PAGSTLVVVDD46 pKa = 5.11GSAVNSTIKK55 pKa = 10.43AGKK58 pKa = 9.86VVLQANDD65 pKa = 4.14PNCDD69 pKa = 3.63PNVAPPCSYY78 pKa = 10.86VLKK81 pKa = 10.54KK82 pKa = 10.62LVIEE86 pKa = 4.26VTDD89 pKa = 4.25FSFKK93 pKa = 11.0DD94 pKa = 3.48EE95 pKa = 4.29NVTNGQFITTGPVNVVDD112 pKa = 4.34DD113 pKa = 4.33GGGIRR118 pKa = 11.84LPAGTPIGFAVTVGDD133 pKa = 4.53DD134 pKa = 3.22RR135 pKa = 11.84AGGIGSVAAPVLIQLDD151 pKa = 3.85VASQQMSLTGSFQGSLEE168 pKa = 4.13GFDD171 pKa = 3.25VAAAVAAQAQSPFVNVPPKK190 pKa = 10.41ISIEE194 pKa = 4.03KK195 pKa = 8.69TLQALEE201 pKa = 4.75CEE203 pKa = 5.06PIPLVASVTDD213 pKa = 3.76ATPAPNDD220 pKa = 3.99DD221 pKa = 3.54VFAPAWAIDD230 pKa = 3.61GVNQGFAPAGFASLAPGAHH249 pKa = 6.09FVQASVQDD257 pKa = 3.67FFGALAVDD265 pKa = 3.84SFVVNVSPDD274 pKa = 3.21EE275 pKa = 4.39APPVFTFVPSDD286 pKa = 3.39IKK288 pKa = 10.27TSSCGGLNIGVATAVDD304 pKa = 3.32NCGEE308 pKa = 4.2VEE310 pKa = 4.41VTSDD314 pKa = 3.81APAKK318 pKa = 9.6FAAGTTVVSWTARR331 pKa = 11.84DD332 pKa = 3.38ARR334 pKa = 11.84GNEE337 pKa = 3.88TVAQQVVTASLKK349 pKa = 10.83DD350 pKa = 3.72DD351 pKa = 4.6ASCCPVGSNVLVGSANNDD369 pKa = 3.32NLVGSSGPDD378 pKa = 3.26CILGLRR384 pKa = 11.84GQDD387 pKa = 3.42QIDD390 pKa = 3.76GGGGDD395 pKa = 4.46DD396 pKa = 4.71VISGGDD402 pKa = 3.26GDD404 pKa = 5.43DD405 pKa = 3.45ILRR408 pKa = 11.84AGAGNDD414 pKa = 4.0LVWAGPGQDD423 pKa = 3.58QLFGGIGDD431 pKa = 4.97DD432 pKa = 4.32LLSGNDD438 pKa = 3.6GDD440 pKa = 5.0DD441 pKa = 3.6LLRR444 pKa = 11.84GEE446 pKa = 5.14GGLDD450 pKa = 3.3TLVGGQGQDD459 pKa = 2.63ILYY462 pKa = 10.75GGDD465 pKa = 4.05DD466 pKa = 3.69NDD468 pKa = 3.97ILTGEE473 pKa = 4.71AGDD476 pKa = 3.95DD477 pKa = 3.62QLFGEE482 pKa = 5.07AGDD485 pKa = 4.49DD486 pKa = 4.0VLDD489 pKa = 5.07GGDD492 pKa = 4.31NNDD495 pKa = 3.32KK496 pKa = 10.01CTGGSGYY503 pKa = 9.05DD504 pKa = 3.38TMLSCTALDD513 pKa = 3.61TLEE516 pKa = 4.79NPGPPSFPGSPSYY529 pKa = 9.94EE530 pKa = 4.06VCEE533 pKa = 4.16CRR535 pKa = 11.84PNKK538 pKa = 10.31CADD541 pKa = 3.47CAAPVATCDD550 pKa = 4.04ASNGCAQIVQCVRR563 pKa = 11.84QAANCNLPHH572 pKa = 6.62EE573 pKa = 4.91CSALCEE579 pKa = 4.28SGRR582 pKa = 11.84TQSAIGAARR591 pKa = 11.84QLASCFGGCC600 pKa = 4.91
Molecular weight: 60.43 kDa
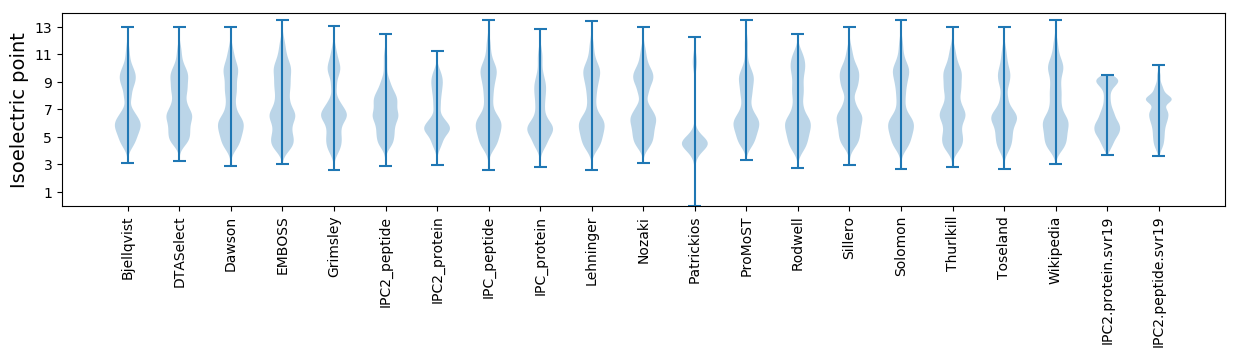
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q5XLE6|A0A4Q5XLE6_9DELT Uncharacterized protein OS=Myxococcales bacterium OX=2026763 GN=EOO73_00325 PE=4 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84HH3 pKa = 5.35GGGWSSRR10 pKa = 11.84EE11 pKa = 3.65RR12 pKa = 11.84HH13 pKa = 5.61LSGHH17 pKa = 6.75AGRR20 pKa = 11.84GRR22 pKa = 11.84AGRR25 pKa = 11.84GRR27 pKa = 11.84AGRR30 pKa = 11.84GRR32 pKa = 11.84AGRR35 pKa = 11.84GRR37 pKa = 11.84AGRR40 pKa = 11.84GRR42 pKa = 11.84AGRR45 pKa = 11.84GRR47 pKa = 11.84AGRR50 pKa = 11.84GRR52 pKa = 11.84AGRR55 pKa = 11.84GRR57 pKa = 11.84AGRR60 pKa = 11.84GRR62 pKa = 11.84AGHH65 pKa = 6.92RR66 pKa = 11.84GRR68 pKa = 11.84GG69 pKa = 3.43
MM1 pKa = 7.55RR2 pKa = 11.84HH3 pKa = 5.35GGGWSSRR10 pKa = 11.84EE11 pKa = 3.65RR12 pKa = 11.84HH13 pKa = 5.61LSGHH17 pKa = 6.75AGRR20 pKa = 11.84GRR22 pKa = 11.84AGRR25 pKa = 11.84GRR27 pKa = 11.84AGRR30 pKa = 11.84GRR32 pKa = 11.84AGRR35 pKa = 11.84GRR37 pKa = 11.84AGRR40 pKa = 11.84GRR42 pKa = 11.84AGRR45 pKa = 11.84GRR47 pKa = 11.84AGRR50 pKa = 11.84GRR52 pKa = 11.84AGRR55 pKa = 11.84GRR57 pKa = 11.84AGRR60 pKa = 11.84GRR62 pKa = 11.84AGHH65 pKa = 6.92RR66 pKa = 11.84GRR68 pKa = 11.84GG69 pKa = 3.43
Molecular weight: 7.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2565783 |
28 |
4227 |
358.8 |
38.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.857 ± 0.042 | 1.189 ± 0.021 |
5.2 ± 0.023 | 6.267 ± 0.029 |
3.29 ± 0.017 | 8.878 ± 0.038 |
1.92 ± 0.014 | 3.231 ± 0.018 |
3.06 ± 0.025 | 10.81 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.662 ± 0.012 | 2.096 ± 0.018 |
5.922 ± 0.028 | 3.244 ± 0.015 |
7.637 ± 0.036 | 6.614 ± 0.026 |
5.066 ± 0.023 | 7.73 ± 0.023 |
1.234 ± 0.011 | 2.095 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
