
Desulfosarcina widdelii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfosarcina
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
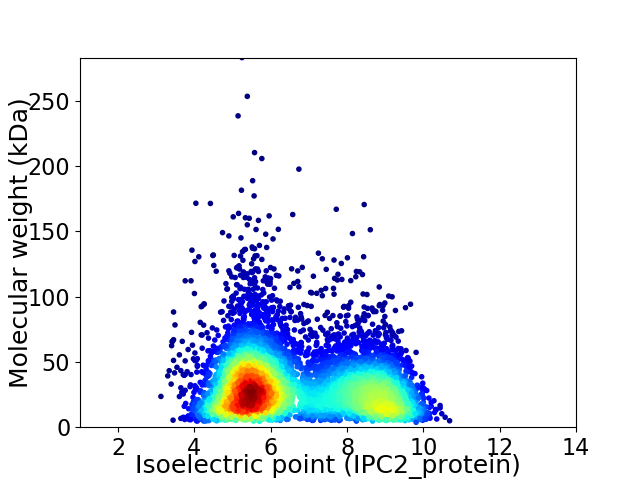
Virtual 2D-PAGE plot for 6679 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5K7Z4E5|A0A5K7Z4E5_9DELT Uncharacterized protein OS=Desulfosarcina widdelii OX=947919 GN=DSCW_32750 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 10.27KK3 pKa = 10.43VSLLIALCSALMIAAPVMALEE24 pKa = 4.66VEE26 pKa = 4.6VSGHH30 pKa = 5.3YY31 pKa = 9.74FVEE34 pKa = 4.99TYY36 pKa = 11.23NNSNSNLSPNDD47 pKa = 3.29ATDD50 pKa = 4.79DD51 pKa = 3.78YY52 pKa = 12.05SKK54 pKa = 10.48MEE56 pKa = 4.23FMAKK60 pKa = 9.66PVFKK64 pKa = 10.01ITDD67 pKa = 4.21NITLTTQFTGLEE79 pKa = 4.02GHH81 pKa = 6.09VWGDD85 pKa = 3.52NPSDD89 pKa = 3.67AGSEE93 pKa = 4.27FDD95 pKa = 5.32DD96 pKa = 5.2DD97 pKa = 4.69YY98 pKa = 12.23NNIEE102 pKa = 3.89WKK104 pKa = 9.87AAYY107 pKa = 7.5MTIKK111 pKa = 9.79TPIGGFIVGRR121 pKa = 11.84YY122 pKa = 9.01IDD124 pKa = 4.44TPWGTDD130 pKa = 3.23LGDD133 pKa = 3.53STASHH138 pKa = 6.96GSNDD142 pKa = 3.51LAKK145 pKa = 10.68DD146 pKa = 3.61RR147 pKa = 11.84IMWVVPVGDD156 pKa = 5.0FISGLVYY163 pKa = 10.49QRR165 pKa = 11.84VGEE168 pKa = 4.08GDD170 pKa = 3.47KK171 pKa = 11.26GVSDD175 pKa = 3.81SDD177 pKa = 3.35KK178 pKa = 11.55DD179 pKa = 3.7NIRR182 pKa = 11.84AYY184 pKa = 10.67GFTAYY189 pKa = 9.53KK190 pKa = 9.26QEE192 pKa = 3.84NWSTGLLVSRR202 pKa = 11.84YY203 pKa = 7.27NLKK206 pKa = 10.89GFVDD210 pKa = 3.9MADD213 pKa = 3.51LRR215 pKa = 11.84ALQAGYY221 pKa = 11.24NNYY224 pKa = 9.25EE225 pKa = 4.22ALNDD229 pKa = 3.89AATLAQATYY238 pKa = 10.43DD239 pKa = 3.37GAYY242 pKa = 8.6AQIYY246 pKa = 10.1AGLAPLLGEE255 pKa = 4.38DD256 pKa = 3.53AAYY259 pKa = 10.21AAIQGEE265 pKa = 4.31LAATNDD271 pKa = 3.47FALGQAVDD279 pKa = 4.04LANLQRR285 pKa = 11.84DD286 pKa = 3.46ASLAAGQAAAAGSLISGGLTNSTAAFWVLDD316 pKa = 4.17PYY318 pKa = 11.35FKK320 pKa = 10.76GTFGPLGIEE329 pKa = 4.3AEE331 pKa = 4.05FLYY334 pKa = 11.01AFGTLEE340 pKa = 4.19LDD342 pKa = 3.54EE343 pKa = 4.75TRR345 pKa = 11.84TDD347 pKa = 3.23VRR349 pKa = 11.84NGSQFDD355 pKa = 3.97EE356 pKa = 4.73LDD358 pKa = 3.99GEE360 pKa = 4.5AMAATIDD367 pKa = 3.87LKK369 pKa = 10.4YY370 pKa = 9.13TIAGFTFNGGYY381 pKa = 8.96TYY383 pKa = 11.22VQGDD387 pKa = 4.12SDD389 pKa = 3.67WADD392 pKa = 3.7DD393 pKa = 3.87EE394 pKa = 4.89FGSIGYY400 pKa = 9.52LEE402 pKa = 3.91QSIDD406 pKa = 3.79LEE408 pKa = 4.57HH409 pKa = 6.89GFLLTSDD416 pKa = 3.67TADD419 pKa = 3.2MEE421 pKa = 4.8GTFGGTDD428 pKa = 3.16ANGIALGNLAGGRR441 pKa = 11.84QTLAGCAGYY450 pKa = 10.96QMYY453 pKa = 9.79WLGAEE458 pKa = 4.16YY459 pKa = 10.3QVLDD463 pKa = 3.78NLKK466 pKa = 9.96IGALFVSSKK475 pKa = 11.29ADD477 pKa = 3.52DD478 pKa = 4.16APYY481 pKa = 11.14LDD483 pKa = 4.41LTSTDD488 pKa = 3.32KK489 pKa = 11.24EE490 pKa = 4.27QWDD493 pKa = 3.98DD494 pKa = 3.92DD495 pKa = 4.0HH496 pKa = 7.85GQEE499 pKa = 4.72YY500 pKa = 10.96DD501 pKa = 4.88LTVEE505 pKa = 4.0WNIMDD510 pKa = 4.37NLTFNGVVAHH520 pKa = 6.76LAAGDD525 pKa = 3.57FWKK528 pKa = 10.95AGDD531 pKa = 4.1PDD533 pKa = 5.41AEE535 pKa = 4.51IEE537 pKa = 4.86DD538 pKa = 3.62NTTFYY543 pKa = 11.13GQLIVEE549 pKa = 4.79FF550 pKa = 4.61
MM1 pKa = 7.52KK2 pKa = 10.27KK3 pKa = 10.43VSLLIALCSALMIAAPVMALEE24 pKa = 4.66VEE26 pKa = 4.6VSGHH30 pKa = 5.3YY31 pKa = 9.74FVEE34 pKa = 4.99TYY36 pKa = 11.23NNSNSNLSPNDD47 pKa = 3.29ATDD50 pKa = 4.79DD51 pKa = 3.78YY52 pKa = 12.05SKK54 pKa = 10.48MEE56 pKa = 4.23FMAKK60 pKa = 9.66PVFKK64 pKa = 10.01ITDD67 pKa = 4.21NITLTTQFTGLEE79 pKa = 4.02GHH81 pKa = 6.09VWGDD85 pKa = 3.52NPSDD89 pKa = 3.67AGSEE93 pKa = 4.27FDD95 pKa = 5.32DD96 pKa = 5.2DD97 pKa = 4.69YY98 pKa = 12.23NNIEE102 pKa = 3.89WKK104 pKa = 9.87AAYY107 pKa = 7.5MTIKK111 pKa = 9.79TPIGGFIVGRR121 pKa = 11.84YY122 pKa = 9.01IDD124 pKa = 4.44TPWGTDD130 pKa = 3.23LGDD133 pKa = 3.53STASHH138 pKa = 6.96GSNDD142 pKa = 3.51LAKK145 pKa = 10.68DD146 pKa = 3.61RR147 pKa = 11.84IMWVVPVGDD156 pKa = 5.0FISGLVYY163 pKa = 10.49QRR165 pKa = 11.84VGEE168 pKa = 4.08GDD170 pKa = 3.47KK171 pKa = 11.26GVSDD175 pKa = 3.81SDD177 pKa = 3.35KK178 pKa = 11.55DD179 pKa = 3.7NIRR182 pKa = 11.84AYY184 pKa = 10.67GFTAYY189 pKa = 9.53KK190 pKa = 9.26QEE192 pKa = 3.84NWSTGLLVSRR202 pKa = 11.84YY203 pKa = 7.27NLKK206 pKa = 10.89GFVDD210 pKa = 3.9MADD213 pKa = 3.51LRR215 pKa = 11.84ALQAGYY221 pKa = 11.24NNYY224 pKa = 9.25EE225 pKa = 4.22ALNDD229 pKa = 3.89AATLAQATYY238 pKa = 10.43DD239 pKa = 3.37GAYY242 pKa = 8.6AQIYY246 pKa = 10.1AGLAPLLGEE255 pKa = 4.38DD256 pKa = 3.53AAYY259 pKa = 10.21AAIQGEE265 pKa = 4.31LAATNDD271 pKa = 3.47FALGQAVDD279 pKa = 4.04LANLQRR285 pKa = 11.84DD286 pKa = 3.46ASLAAGQAAAAGSLISGGLTNSTAAFWVLDD316 pKa = 4.17PYY318 pKa = 11.35FKK320 pKa = 10.76GTFGPLGIEE329 pKa = 4.3AEE331 pKa = 4.05FLYY334 pKa = 11.01AFGTLEE340 pKa = 4.19LDD342 pKa = 3.54EE343 pKa = 4.75TRR345 pKa = 11.84TDD347 pKa = 3.23VRR349 pKa = 11.84NGSQFDD355 pKa = 3.97EE356 pKa = 4.73LDD358 pKa = 3.99GEE360 pKa = 4.5AMAATIDD367 pKa = 3.87LKK369 pKa = 10.4YY370 pKa = 9.13TIAGFTFNGGYY381 pKa = 8.96TYY383 pKa = 11.22VQGDD387 pKa = 4.12SDD389 pKa = 3.67WADD392 pKa = 3.7DD393 pKa = 3.87EE394 pKa = 4.89FGSIGYY400 pKa = 9.52LEE402 pKa = 3.91QSIDD406 pKa = 3.79LEE408 pKa = 4.57HH409 pKa = 6.89GFLLTSDD416 pKa = 3.67TADD419 pKa = 3.2MEE421 pKa = 4.8GTFGGTDD428 pKa = 3.16ANGIALGNLAGGRR441 pKa = 11.84QTLAGCAGYY450 pKa = 10.96QMYY453 pKa = 9.79WLGAEE458 pKa = 4.16YY459 pKa = 10.3QVLDD463 pKa = 3.78NLKK466 pKa = 9.96IGALFVSSKK475 pKa = 11.29ADD477 pKa = 3.52DD478 pKa = 4.16APYY481 pKa = 11.14LDD483 pKa = 4.41LTSTDD488 pKa = 3.32KK489 pKa = 11.24EE490 pKa = 4.27QWDD493 pKa = 3.98DD494 pKa = 3.92DD495 pKa = 4.0HH496 pKa = 7.85GQEE499 pKa = 4.72YY500 pKa = 10.96DD501 pKa = 4.88LTVEE505 pKa = 4.0WNIMDD510 pKa = 4.37NLTFNGVVAHH520 pKa = 6.76LAAGDD525 pKa = 3.57FWKK528 pKa = 10.95AGDD531 pKa = 4.1PDD533 pKa = 5.41AEE535 pKa = 4.51IEE537 pKa = 4.86DD538 pKa = 3.62NTTFYY543 pKa = 11.13GQLIVEE549 pKa = 4.79FF550 pKa = 4.61
Molecular weight: 59.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5K7ZFJ8|A0A5K7ZFJ8_9DELT 2-dehydropantoate 2-reductase OS=Desulfosarcina widdelii OX=947919 GN=DSCW_59100 PE=3 SV=1
MM1 pKa = 7.5LLAGALAIPFHH12 pKa = 6.68FEE14 pKa = 4.36SPSMWYY20 pKa = 10.28KK21 pKa = 10.75FGAAKK26 pKa = 10.21ISLRR30 pKa = 11.84AGKK33 pKa = 8.61MLGMAAGLMILLQLPLAGRR52 pKa = 11.84LKK54 pKa = 10.83LLDD57 pKa = 5.09RR58 pKa = 11.84IFSLPGLIRR67 pKa = 11.84QHH69 pKa = 6.35RR70 pKa = 11.84WHH72 pKa = 6.83AWAIVIMALIHH83 pKa = 5.75PLCVLLAEE91 pKa = 4.95GKK93 pKa = 10.45LLIPLEE99 pKa = 3.98MRR101 pKa = 11.84YY102 pKa = 8.33WPEE105 pKa = 3.38WVGVGLLVVLLAQFACTQWRR125 pKa = 11.84RR126 pKa = 11.84PLKK129 pKa = 10.31IAFHH133 pKa = 6.52HH134 pKa = 6.77WLTGHH139 pKa = 7.38RR140 pKa = 11.84IAGLITAALLIVHH153 pKa = 5.83VRR155 pKa = 11.84YY156 pKa = 10.31VSEE159 pKa = 4.36TFTDD163 pKa = 3.68NPPPRR168 pKa = 11.84LAVLSAAGVFTLFWLWIRR186 pKa = 11.84STWLRR191 pKa = 11.84TRR193 pKa = 11.84RR194 pKa = 11.84TPFSVTRR201 pKa = 11.84VEE203 pKa = 4.54NIGRR207 pKa = 11.84EE208 pKa = 4.4CTCVEE213 pKa = 3.97VAPEE217 pKa = 3.9KK218 pKa = 10.14GAAMAYY224 pKa = 10.3FPGQFAFVSFTSQILSRR241 pKa = 11.84EE242 pKa = 3.99PHH244 pKa = 6.73PFTLASTPSRR254 pKa = 11.84PGTIQFAIRR263 pKa = 11.84HH264 pKa = 5.73CGDD267 pKa = 2.63WTRR270 pKa = 11.84QVEE273 pKa = 4.27TLAIGDD279 pKa = 3.73RR280 pKa = 11.84AYY282 pKa = 11.13LQGPFGRR289 pKa = 11.84FGHH292 pKa = 7.29LLQASGQQEE301 pKa = 4.94LIMIAGGIGITPMLSMLRR319 pKa = 11.84FMADD323 pKa = 2.59HH324 pKa = 7.18GDD326 pKa = 3.65PRR328 pKa = 11.84PITLIWSNRR337 pKa = 11.84TRR339 pKa = 11.84TQLVFTEE346 pKa = 4.81EE347 pKa = 4.12FQHH350 pKa = 7.19LSAKK354 pKa = 9.2LTGLRR359 pKa = 11.84RR360 pKa = 11.84VPIFTEE366 pKa = 4.22NADD369 pKa = 3.37SGTPSGRR376 pKa = 11.84LNRR379 pKa = 11.84IMLQSLLSGCSRR391 pKa = 11.84GSAVFLCGPPSMMTALTTEE410 pKa = 4.68LKK412 pKa = 10.89ALGFSARR419 pKa = 11.84LIITEE424 pKa = 4.11TFGFF428 pKa = 4.04
MM1 pKa = 7.5LLAGALAIPFHH12 pKa = 6.68FEE14 pKa = 4.36SPSMWYY20 pKa = 10.28KK21 pKa = 10.75FGAAKK26 pKa = 10.21ISLRR30 pKa = 11.84AGKK33 pKa = 8.61MLGMAAGLMILLQLPLAGRR52 pKa = 11.84LKK54 pKa = 10.83LLDD57 pKa = 5.09RR58 pKa = 11.84IFSLPGLIRR67 pKa = 11.84QHH69 pKa = 6.35RR70 pKa = 11.84WHH72 pKa = 6.83AWAIVIMALIHH83 pKa = 5.75PLCVLLAEE91 pKa = 4.95GKK93 pKa = 10.45LLIPLEE99 pKa = 3.98MRR101 pKa = 11.84YY102 pKa = 8.33WPEE105 pKa = 3.38WVGVGLLVVLLAQFACTQWRR125 pKa = 11.84RR126 pKa = 11.84PLKK129 pKa = 10.31IAFHH133 pKa = 6.52HH134 pKa = 6.77WLTGHH139 pKa = 7.38RR140 pKa = 11.84IAGLITAALLIVHH153 pKa = 5.83VRR155 pKa = 11.84YY156 pKa = 10.31VSEE159 pKa = 4.36TFTDD163 pKa = 3.68NPPPRR168 pKa = 11.84LAVLSAAGVFTLFWLWIRR186 pKa = 11.84STWLRR191 pKa = 11.84TRR193 pKa = 11.84RR194 pKa = 11.84TPFSVTRR201 pKa = 11.84VEE203 pKa = 4.54NIGRR207 pKa = 11.84EE208 pKa = 4.4CTCVEE213 pKa = 3.97VAPEE217 pKa = 3.9KK218 pKa = 10.14GAAMAYY224 pKa = 10.3FPGQFAFVSFTSQILSRR241 pKa = 11.84EE242 pKa = 3.99PHH244 pKa = 6.73PFTLASTPSRR254 pKa = 11.84PGTIQFAIRR263 pKa = 11.84HH264 pKa = 5.73CGDD267 pKa = 2.63WTRR270 pKa = 11.84QVEE273 pKa = 4.27TLAIGDD279 pKa = 3.73RR280 pKa = 11.84AYY282 pKa = 11.13LQGPFGRR289 pKa = 11.84FGHH292 pKa = 7.29LLQASGQQEE301 pKa = 4.94LIMIAGGIGITPMLSMLRR319 pKa = 11.84FMADD323 pKa = 2.59HH324 pKa = 7.18GDD326 pKa = 3.65PRR328 pKa = 11.84PITLIWSNRR337 pKa = 11.84TRR339 pKa = 11.84TQLVFTEE346 pKa = 4.81EE347 pKa = 4.12FQHH350 pKa = 7.19LSAKK354 pKa = 9.2LTGLRR359 pKa = 11.84RR360 pKa = 11.84VPIFTEE366 pKa = 4.22NADD369 pKa = 3.37SGTPSGRR376 pKa = 11.84LNRR379 pKa = 11.84IMLQSLLSGCSRR391 pKa = 11.84GSAVFLCGPPSMMTALTTEE410 pKa = 4.68LKK412 pKa = 10.89ALGFSARR419 pKa = 11.84LIITEE424 pKa = 4.11TFGFF428 pKa = 4.04
Molecular weight: 47.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2063912 |
39 |
2590 |
309.0 |
34.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.09 ± 0.036 | 1.297 ± 0.014 |
5.831 ± 0.025 | 6.228 ± 0.026 |
4.189 ± 0.02 | 7.583 ± 0.026 |
2.07 ± 0.013 | 6.528 ± 0.028 |
5.258 ± 0.028 | 9.661 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.814 ± 0.015 | 3.436 ± 0.016 |
4.514 ± 0.02 | 3.359 ± 0.016 |
6.202 ± 0.026 | 5.572 ± 0.02 |
5.241 ± 0.017 | 7.108 ± 0.025 |
1.16 ± 0.014 | 2.858 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
