
Beihai tombus-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
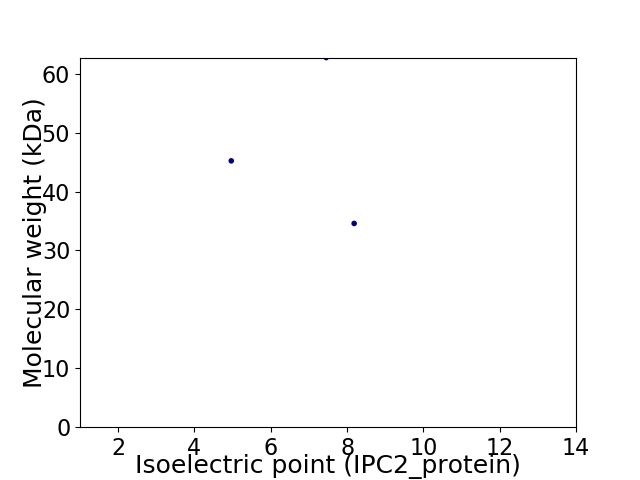
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFW4|A0A1L3KFW4_9VIRU Capsid protein OS=Beihai tombus-like virus 3 OX=1922724 PE=3 SV=1
MM1 pKa = 7.79RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 8.07TNNARR9 pKa = 11.84RR10 pKa = 11.84VSAPIANGLRR20 pKa = 11.84TSGGRR25 pKa = 11.84PQMTPAGAGSTRR37 pKa = 11.84VRR39 pKa = 11.84HH40 pKa = 6.01RR41 pKa = 11.84EE42 pKa = 3.48FVQNITSDD50 pKa = 3.4HH51 pKa = 5.87TFGSEE56 pKa = 4.42LLALGINVGDD66 pKa = 3.84TKK68 pKa = 10.68TFPWLSKK75 pKa = 9.83IANGYY80 pKa = 8.17EE81 pKa = 4.3RR82 pKa = 11.84YY83 pKa = 9.06CVHH86 pKa = 6.91SMTISYY92 pKa = 10.38EE93 pKa = 4.08PFVSTTEE100 pKa = 3.74TGAILMLADD109 pKa = 4.65YY110 pKa = 10.28DD111 pKa = 4.49PNDD114 pKa = 4.32PPPTSKK120 pKa = 11.34SNMLNSMGASRR131 pKa = 11.84SAVWMNTTMPLDD143 pKa = 4.0RR144 pKa = 11.84KK145 pKa = 9.58EE146 pKa = 4.55LSYY149 pKa = 11.46DD150 pKa = 2.96DD151 pKa = 4.48HH152 pKa = 8.62LFVRR156 pKa = 11.84HH157 pKa = 4.97QTRR160 pKa = 11.84EE161 pKa = 4.01TLTEE165 pKa = 3.77NLKK168 pKa = 10.89LYY170 pKa = 10.68DD171 pKa = 3.56VGTVFVGVTDD181 pKa = 3.97NEE183 pKa = 4.46PVSGTKK189 pKa = 9.84QYY191 pKa = 11.11GEE193 pKa = 3.71VWITYY198 pKa = 9.92DD199 pKa = 3.29VTLMVPAFHH208 pKa = 6.79TSNPNSAEE216 pKa = 4.09TKK218 pKa = 9.92VNGAIAPDD226 pKa = 3.95FLGGIQSSNRR236 pKa = 11.84AAMKK240 pKa = 10.54DD241 pKa = 3.33GSAVNFSTAHH251 pKa = 6.05NGDD254 pKa = 3.77DD255 pKa = 3.39QVAITFHH262 pKa = 6.55EE263 pKa = 4.85PFTGLLYY270 pKa = 10.06MEE272 pKa = 4.29QSGYY276 pKa = 11.54ALDD279 pKa = 4.46DD280 pKa = 3.67TTHH283 pKa = 5.6VQLEE287 pKa = 4.68PEE289 pKa = 4.19TSPTDD294 pKa = 3.21GWISKK299 pKa = 9.15LAKK302 pKa = 10.32LGGITADD309 pKa = 3.75YY310 pKa = 10.08FFQEE314 pKa = 4.38NTWKK318 pKa = 10.93YY319 pKa = 10.34LMEE322 pKa = 4.28VVADD326 pKa = 3.93AGEE329 pKa = 4.54SIVFSALGVGAADD342 pKa = 3.03IATWIGEE349 pKa = 4.54VAWLLAPYY357 pKa = 10.77AEE359 pKa = 4.26TLMFAPLLVLNKK371 pKa = 10.14ASAIDD376 pKa = 3.48VRR378 pKa = 11.84ARR380 pKa = 11.84LQARR384 pKa = 11.84SLNGILPANEE394 pKa = 3.77VLPRR398 pKa = 11.84LDD400 pKa = 3.04WVLQQSNQDD409 pKa = 3.48PEE411 pKa = 4.18EE412 pKa = 4.0
MM1 pKa = 7.79RR2 pKa = 11.84RR3 pKa = 11.84KK4 pKa = 8.07TNNARR9 pKa = 11.84RR10 pKa = 11.84VSAPIANGLRR20 pKa = 11.84TSGGRR25 pKa = 11.84PQMTPAGAGSTRR37 pKa = 11.84VRR39 pKa = 11.84HH40 pKa = 6.01RR41 pKa = 11.84EE42 pKa = 3.48FVQNITSDD50 pKa = 3.4HH51 pKa = 5.87TFGSEE56 pKa = 4.42LLALGINVGDD66 pKa = 3.84TKK68 pKa = 10.68TFPWLSKK75 pKa = 9.83IANGYY80 pKa = 8.17EE81 pKa = 4.3RR82 pKa = 11.84YY83 pKa = 9.06CVHH86 pKa = 6.91SMTISYY92 pKa = 10.38EE93 pKa = 4.08PFVSTTEE100 pKa = 3.74TGAILMLADD109 pKa = 4.65YY110 pKa = 10.28DD111 pKa = 4.49PNDD114 pKa = 4.32PPPTSKK120 pKa = 11.34SNMLNSMGASRR131 pKa = 11.84SAVWMNTTMPLDD143 pKa = 4.0RR144 pKa = 11.84KK145 pKa = 9.58EE146 pKa = 4.55LSYY149 pKa = 11.46DD150 pKa = 2.96DD151 pKa = 4.48HH152 pKa = 8.62LFVRR156 pKa = 11.84HH157 pKa = 4.97QTRR160 pKa = 11.84EE161 pKa = 4.01TLTEE165 pKa = 3.77NLKK168 pKa = 10.89LYY170 pKa = 10.68DD171 pKa = 3.56VGTVFVGVTDD181 pKa = 3.97NEE183 pKa = 4.46PVSGTKK189 pKa = 9.84QYY191 pKa = 11.11GEE193 pKa = 3.71VWITYY198 pKa = 9.92DD199 pKa = 3.29VTLMVPAFHH208 pKa = 6.79TSNPNSAEE216 pKa = 4.09TKK218 pKa = 9.92VNGAIAPDD226 pKa = 3.95FLGGIQSSNRR236 pKa = 11.84AAMKK240 pKa = 10.54DD241 pKa = 3.33GSAVNFSTAHH251 pKa = 6.05NGDD254 pKa = 3.77DD255 pKa = 3.39QVAITFHH262 pKa = 6.55EE263 pKa = 4.85PFTGLLYY270 pKa = 10.06MEE272 pKa = 4.29QSGYY276 pKa = 11.54ALDD279 pKa = 4.46DD280 pKa = 3.67TTHH283 pKa = 5.6VQLEE287 pKa = 4.68PEE289 pKa = 4.19TSPTDD294 pKa = 3.21GWISKK299 pKa = 9.15LAKK302 pKa = 10.32LGGITADD309 pKa = 3.75YY310 pKa = 10.08FFQEE314 pKa = 4.38NTWKK318 pKa = 10.93YY319 pKa = 10.34LMEE322 pKa = 4.28VVADD326 pKa = 3.93AGEE329 pKa = 4.54SIVFSALGVGAADD342 pKa = 3.03IATWIGEE349 pKa = 4.54VAWLLAPYY357 pKa = 10.77AEE359 pKa = 4.26TLMFAPLLVLNKK371 pKa = 10.14ASAIDD376 pKa = 3.48VRR378 pKa = 11.84ARR380 pKa = 11.84LQARR384 pKa = 11.84SLNGILPANEE394 pKa = 3.77VLPRR398 pKa = 11.84LDD400 pKa = 3.04WVLQQSNQDD409 pKa = 3.48PEE411 pKa = 4.18EE412 pKa = 4.0
Molecular weight: 45.21 kDa
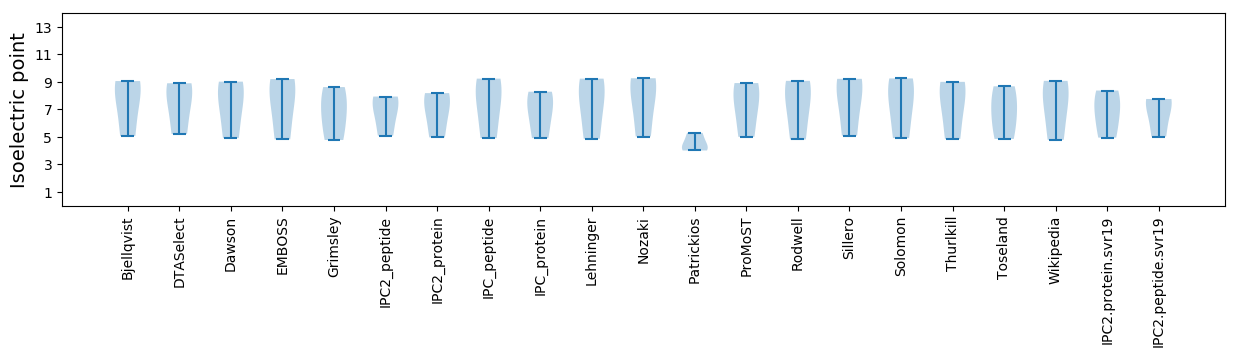
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFW4|A0A1L3KFW4_9VIRU Capsid protein OS=Beihai tombus-like virus 3 OX=1922724 PE=3 SV=1
MM1 pKa = 7.59RR2 pKa = 11.84ASLFPKK8 pKa = 9.0RR9 pKa = 11.84TTGEE13 pKa = 4.02VVGLVDD19 pKa = 4.92GPVPTPQEE27 pKa = 3.89VRR29 pKa = 11.84GGPFSWDD36 pKa = 3.62KK37 pKa = 11.48AWLRR41 pKa = 11.84CMNTLKK47 pKa = 10.78QNLQSEE53 pKa = 4.65IVGCVNHH60 pKa = 5.96NHH62 pKa = 5.67VDD64 pKa = 3.7PRR66 pKa = 11.84PEE68 pKa = 3.34WEE70 pKa = 3.87IAVTEE75 pKa = 4.39GLASTGNVIINNTIRR90 pKa = 11.84STGFNVSVTAGGAVGTVGVICGMGVVVSYY119 pKa = 11.1APLVAGVAATWAVSALRR136 pKa = 11.84RR137 pKa = 11.84EE138 pKa = 4.04RR139 pKa = 11.84SHH141 pKa = 7.46AVDD144 pKa = 3.28VTLSVLNDD152 pKa = 3.52DD153 pKa = 5.11VRR155 pKa = 11.84IGDD158 pKa = 4.35LKK160 pKa = 10.9AAGGKK165 pKa = 9.43EE166 pKa = 3.93EE167 pKa = 5.77AGGEE171 pKa = 3.92DD172 pKa = 3.44QAAVDD177 pKa = 4.56SEE179 pKa = 4.79DD180 pKa = 3.7NPAEE184 pKa = 4.11STRR187 pKa = 11.84EE188 pKa = 3.59RR189 pKa = 11.84RR190 pKa = 11.84EE191 pKa = 3.58FEE193 pKa = 3.76VRR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84KK198 pKa = 9.86PFFGPQPRR206 pKa = 11.84NHH208 pKa = 5.57GRR210 pKa = 11.84IPVMAGEE217 pKa = 4.24VANLLKK223 pKa = 10.53LRR225 pKa = 11.84HH226 pKa = 6.07IGLRR230 pKa = 11.84DD231 pKa = 3.39TPEE234 pKa = 3.39NRR236 pKa = 11.84FLIRR240 pKa = 11.84ADD242 pKa = 3.04AGKK245 pKa = 10.07RR246 pKa = 11.84AEE248 pKa = 3.84ALRR251 pKa = 11.84RR252 pKa = 11.84EE253 pKa = 4.84GEE255 pKa = 4.31RR256 pKa = 11.84PWNTVRR262 pKa = 11.84NAEE265 pKa = 4.19LLSIAMHH272 pKa = 6.45ASEE275 pKa = 5.21MYY277 pKa = 9.72WILSRR282 pKa = 11.84DD283 pKa = 3.36EE284 pKa = 4.51EE285 pKa = 4.91YY286 pKa = 11.18VGEE289 pKa = 4.52LYY291 pKa = 10.68SEE293 pKa = 4.22SLLRR297 pKa = 11.84GLRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84NRR305 pKa = 11.84WVSSSVSSKK314 pKa = 11.17
MM1 pKa = 7.59RR2 pKa = 11.84ASLFPKK8 pKa = 9.0RR9 pKa = 11.84TTGEE13 pKa = 4.02VVGLVDD19 pKa = 4.92GPVPTPQEE27 pKa = 3.89VRR29 pKa = 11.84GGPFSWDD36 pKa = 3.62KK37 pKa = 11.48AWLRR41 pKa = 11.84CMNTLKK47 pKa = 10.78QNLQSEE53 pKa = 4.65IVGCVNHH60 pKa = 5.96NHH62 pKa = 5.67VDD64 pKa = 3.7PRR66 pKa = 11.84PEE68 pKa = 3.34WEE70 pKa = 3.87IAVTEE75 pKa = 4.39GLASTGNVIINNTIRR90 pKa = 11.84STGFNVSVTAGGAVGTVGVICGMGVVVSYY119 pKa = 11.1APLVAGVAATWAVSALRR136 pKa = 11.84RR137 pKa = 11.84EE138 pKa = 4.04RR139 pKa = 11.84SHH141 pKa = 7.46AVDD144 pKa = 3.28VTLSVLNDD152 pKa = 3.52DD153 pKa = 5.11VRR155 pKa = 11.84IGDD158 pKa = 4.35LKK160 pKa = 10.9AAGGKK165 pKa = 9.43EE166 pKa = 3.93EE167 pKa = 5.77AGGEE171 pKa = 3.92DD172 pKa = 3.44QAAVDD177 pKa = 4.56SEE179 pKa = 4.79DD180 pKa = 3.7NPAEE184 pKa = 4.11STRR187 pKa = 11.84EE188 pKa = 3.59RR189 pKa = 11.84RR190 pKa = 11.84EE191 pKa = 3.58FEE193 pKa = 3.76VRR195 pKa = 11.84RR196 pKa = 11.84RR197 pKa = 11.84KK198 pKa = 9.86PFFGPQPRR206 pKa = 11.84NHH208 pKa = 5.57GRR210 pKa = 11.84IPVMAGEE217 pKa = 4.24VANLLKK223 pKa = 10.53LRR225 pKa = 11.84HH226 pKa = 6.07IGLRR230 pKa = 11.84DD231 pKa = 3.39TPEE234 pKa = 3.39NRR236 pKa = 11.84FLIRR240 pKa = 11.84ADD242 pKa = 3.04AGKK245 pKa = 10.07RR246 pKa = 11.84AEE248 pKa = 3.84ALRR251 pKa = 11.84RR252 pKa = 11.84EE253 pKa = 4.84GEE255 pKa = 4.31RR256 pKa = 11.84PWNTVRR262 pKa = 11.84NAEE265 pKa = 4.19LLSIAMHH272 pKa = 6.45ASEE275 pKa = 5.21MYY277 pKa = 9.72WILSRR282 pKa = 11.84DD283 pKa = 3.36EE284 pKa = 4.51EE285 pKa = 4.91YY286 pKa = 11.18VGEE289 pKa = 4.52LYY291 pKa = 10.68SEE293 pKa = 4.22SLLRR297 pKa = 11.84GLRR300 pKa = 11.84RR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84NRR305 pKa = 11.84WVSSSVSSKK314 pKa = 11.17
Molecular weight: 34.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1270 |
314 |
544 |
423.3 |
47.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.165 ± 1.394 | 1.339 ± 0.587 |
5.827 ± 0.659 | 6.929 ± 0.612 |
3.858 ± 0.687 | 7.953 ± 0.458 |
2.047 ± 0.062 | 4.646 ± 0.479 |
4.803 ± 1.288 | 8.189 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.992 ± 0.395 | 4.409 ± 0.702 |
5.039 ± 0.131 | 2.598 ± 0.362 |
7.48 ± 1.39 | 7.008 ± 0.126 |
5.197 ± 1.568 | 7.323 ± 1.193 |
1.732 ± 0.252 | 3.465 ± 0.928 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
