
Desertihabitans brevis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Desertihabitans
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
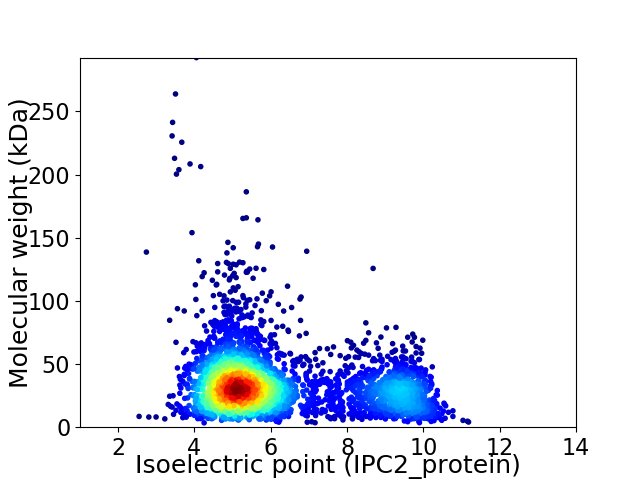
Virtual 2D-PAGE plot for 3682 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A367YYA9|A0A367YYA9_9ACTN DNA-(apurinic or apyrimidinic site) lyase OS=Desertihabitans brevis OX=2268447 GN=DT076_08310 PE=3 SV=1
MM1 pKa = 7.76ALTACGTRR9 pKa = 11.84AEE11 pKa = 4.36EE12 pKa = 4.63GGGGAEE18 pKa = 4.39GGAGGGGGEE27 pKa = 4.57TKK29 pKa = 9.84TAVIGVVAPLSGDD42 pKa = 3.91LAPLGLGIQHH52 pKa = 5.84SVEE55 pKa = 4.26LAVQQANEE63 pKa = 4.15SGAIPGWTLEE73 pKa = 4.09VQPEE77 pKa = 4.04DD78 pKa = 4.06DD79 pKa = 3.7QATPDD84 pKa = 3.46VGRR87 pKa = 11.84NAATSLSSDD96 pKa = 3.3ADD98 pKa = 3.72VVAMVGPLNSSVGQAVQPVFDD119 pKa = 4.21AAGILQVSPANTNPSLTRR137 pKa = 11.84GANFDD142 pKa = 3.78TAPEE146 pKa = 4.14RR147 pKa = 11.84TYY149 pKa = 9.06PTYY152 pKa = 10.77FRR154 pKa = 11.84TCTTDD159 pKa = 3.68SIQGPFAARR168 pKa = 11.84YY169 pKa = 8.76LYY171 pKa = 7.87EE172 pKa = 3.93TAGITRR178 pKa = 11.84IATVHH183 pKa = 6.48DD184 pKa = 4.01KK185 pKa = 10.79KK186 pKa = 11.08AYY188 pKa = 9.12GQGLVEE194 pKa = 4.32AFSAEE199 pKa = 3.98FEE201 pKa = 4.33EE202 pKa = 5.12LGGEE206 pKa = 3.96IVAAEE211 pKa = 4.44TINPEE216 pKa = 4.18DD217 pKa = 4.4DD218 pKa = 3.56NFSAVITSVGSGDD231 pKa = 4.16PEE233 pKa = 3.64ALYY236 pKa = 11.11YY237 pKa = 10.59GGEE240 pKa = 4.23YY241 pKa = 9.09PQSGPLSQQMKK252 pKa = 10.23AAGLDD257 pKa = 3.38IPLMGGDD264 pKa = 3.66GMYY267 pKa = 10.35SPEE270 pKa = 4.3YY271 pKa = 10.16IEE273 pKa = 5.23LAGEE277 pKa = 3.98QSEE280 pKa = 4.86GDD282 pKa = 3.48LATSVGAPVEE292 pKa = 4.39DD293 pKa = 4.9LPSAQQFVTDD303 pKa = 3.71YY304 pKa = 9.88EE305 pKa = 4.35AAGFPDD311 pKa = 4.53PYY313 pKa = 9.75EE314 pKa = 4.59AYY316 pKa = 10.19GAYY319 pKa = 10.2SYY321 pKa = 11.33DD322 pKa = 3.3AAQAVIEE329 pKa = 4.07ALKK332 pKa = 10.89VSLPEE337 pKa = 4.8AEE339 pKa = 4.22TAEE342 pKa = 4.52DD343 pKa = 3.39ARR345 pKa = 11.84EE346 pKa = 4.12ATVEE350 pKa = 3.87AMASVSFEE358 pKa = 3.98GATGPVAFDD367 pKa = 3.34EE368 pKa = 5.33FGDD371 pKa = 3.55NTTRR375 pKa = 11.84ILTAYY380 pKa = 9.08KK381 pKa = 10.37VEE383 pKa = 4.25GGAWVADD390 pKa = 3.57QTDD393 pKa = 3.69EE394 pKa = 5.47FEE396 pKa = 4.58DD397 pKa = 3.64
MM1 pKa = 7.76ALTACGTRR9 pKa = 11.84AEE11 pKa = 4.36EE12 pKa = 4.63GGGGAEE18 pKa = 4.39GGAGGGGGEE27 pKa = 4.57TKK29 pKa = 9.84TAVIGVVAPLSGDD42 pKa = 3.91LAPLGLGIQHH52 pKa = 5.84SVEE55 pKa = 4.26LAVQQANEE63 pKa = 4.15SGAIPGWTLEE73 pKa = 4.09VQPEE77 pKa = 4.04DD78 pKa = 4.06DD79 pKa = 3.7QATPDD84 pKa = 3.46VGRR87 pKa = 11.84NAATSLSSDD96 pKa = 3.3ADD98 pKa = 3.72VVAMVGPLNSSVGQAVQPVFDD119 pKa = 4.21AAGILQVSPANTNPSLTRR137 pKa = 11.84GANFDD142 pKa = 3.78TAPEE146 pKa = 4.14RR147 pKa = 11.84TYY149 pKa = 9.06PTYY152 pKa = 10.77FRR154 pKa = 11.84TCTTDD159 pKa = 3.68SIQGPFAARR168 pKa = 11.84YY169 pKa = 8.76LYY171 pKa = 7.87EE172 pKa = 3.93TAGITRR178 pKa = 11.84IATVHH183 pKa = 6.48DD184 pKa = 4.01KK185 pKa = 10.79KK186 pKa = 11.08AYY188 pKa = 9.12GQGLVEE194 pKa = 4.32AFSAEE199 pKa = 3.98FEE201 pKa = 4.33EE202 pKa = 5.12LGGEE206 pKa = 3.96IVAAEE211 pKa = 4.44TINPEE216 pKa = 4.18DD217 pKa = 4.4DD218 pKa = 3.56NFSAVITSVGSGDD231 pKa = 4.16PEE233 pKa = 3.64ALYY236 pKa = 11.11YY237 pKa = 10.59GGEE240 pKa = 4.23YY241 pKa = 9.09PQSGPLSQQMKK252 pKa = 10.23AAGLDD257 pKa = 3.38IPLMGGDD264 pKa = 3.66GMYY267 pKa = 10.35SPEE270 pKa = 4.3YY271 pKa = 10.16IEE273 pKa = 5.23LAGEE277 pKa = 3.98QSEE280 pKa = 4.86GDD282 pKa = 3.48LATSVGAPVEE292 pKa = 4.39DD293 pKa = 4.9LPSAQQFVTDD303 pKa = 3.71YY304 pKa = 9.88EE305 pKa = 4.35AAGFPDD311 pKa = 4.53PYY313 pKa = 9.75EE314 pKa = 4.59AYY316 pKa = 10.19GAYY319 pKa = 10.2SYY321 pKa = 11.33DD322 pKa = 3.3AAQAVIEE329 pKa = 4.07ALKK332 pKa = 10.89VSLPEE337 pKa = 4.8AEE339 pKa = 4.22TAEE342 pKa = 4.52DD343 pKa = 3.39ARR345 pKa = 11.84EE346 pKa = 4.12ATVEE350 pKa = 3.87AMASVSFEE358 pKa = 3.98GATGPVAFDD367 pKa = 3.34EE368 pKa = 5.33FGDD371 pKa = 3.55NTTRR375 pKa = 11.84ILTAYY380 pKa = 9.08KK381 pKa = 10.37VEE383 pKa = 4.25GGAWVADD390 pKa = 3.57QTDD393 pKa = 3.69EE394 pKa = 5.47FEE396 pKa = 4.58DD397 pKa = 3.64
Molecular weight: 40.94 kDa
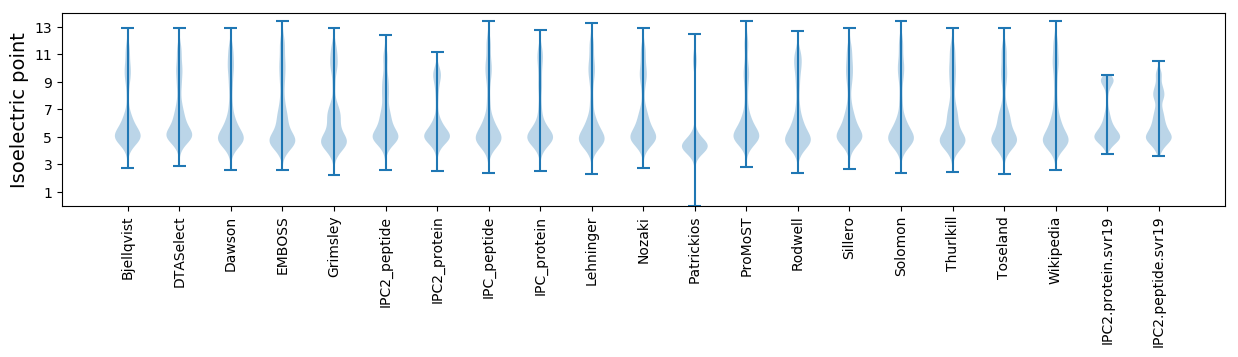
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A367YR83|A0A367YR83_9ACTN ABC transporter permease OS=Desertihabitans brevis OX=2268447 GN=DT076_16485 PE=3 SV=1
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.65KK12 pKa = 10.17IPGAQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 8.79RR31 pKa = 11.84NPRR34 pKa = 11.84FKK36 pKa = 10.78ARR38 pKa = 11.84QGG40 pKa = 3.28
MM1 pKa = 7.61KK2 pKa = 10.31VRR4 pKa = 11.84NSLRR8 pKa = 11.84ALKK11 pKa = 9.65KK12 pKa = 10.17IPGAQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 8.79RR31 pKa = 11.84NPRR34 pKa = 11.84FKK36 pKa = 10.78ARR38 pKa = 11.84QGG40 pKa = 3.28
Molecular weight: 4.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212445 |
31 |
2815 |
329.3 |
35.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.039 ± 0.05 | 0.711 ± 0.011 |
6.201 ± 0.037 | 5.978 ± 0.036 |
2.642 ± 0.026 | 9.343 ± 0.038 |
2.158 ± 0.022 | 2.773 ± 0.033 |
1.294 ± 0.021 | 10.737 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.64 ± 0.017 | 1.545 ± 0.023 |
6.164 ± 0.037 | 3.024 ± 0.022 |
8.277 ± 0.055 | 5.092 ± 0.03 |
6.188 ± 0.036 | 9.712 ± 0.041 |
1.642 ± 0.017 | 1.84 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
