
Pythium brassicum
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Pythiales; Pythiaceae; Pythium
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
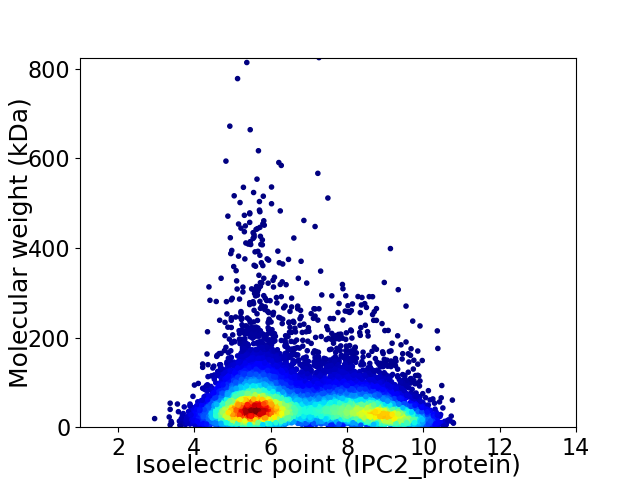
Virtual 2D-PAGE plot for 12998 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5D6XSK8|A0A5D6XSK8_9STRA Uncharacterized protein OS=Pythium brassicum OX=1485010 GN=PybrP1_010047 PE=4 SV=1
MM1 pKa = 7.4TKK3 pKa = 10.1SLHH6 pKa = 5.76LSXVAVTAAAVALLVDD22 pKa = 3.73SGIDD26 pKa = 3.05RR27 pKa = 11.84FGSVGVFAQTTGTAAATVAICRR49 pKa = 11.84APASSLTDD57 pKa = 3.3YY58 pKa = 11.35GVGIVSDD65 pKa = 4.48PACATPSSAASSSCIDD81 pKa = 3.63DD82 pKa = 3.14QCRR85 pKa = 11.84FCQVFRR91 pKa = 11.84TDD93 pKa = 3.2LSKK96 pKa = 11.21GYY98 pKa = 9.79LPCTTPPVLVSGGAGVPPSEE118 pKa = 4.57TPPSPSVPPTTLVPTLASALDD139 pKa = 3.87PTPTAPAAVTFEE151 pKa = 4.29PTVVAPAPASVSMTPVPSAVGIALGAVPATAPPISPLLIDD191 pKa = 4.63CSAEE195 pKa = 3.87LSLEE199 pKa = 3.9AMAAGVYY206 pKa = 10.15AVLDD210 pKa = 4.3PSCQADD216 pKa = 3.71DD217 pKa = 4.54DD218 pKa = 4.49AAQASCQDD226 pKa = 3.64EE227 pKa = 4.56FCRR230 pKa = 11.84LCRR233 pKa = 11.84VGEE236 pKa = 3.95SDD238 pKa = 3.97YY239 pKa = 11.81AANTDD244 pKa = 4.63DD245 pKa = 5.38AGADD249 pKa = 3.82PGTDD253 pKa = 3.58DD254 pKa = 4.57IGANSSADD262 pKa = 3.64NPGTDD267 pKa = 4.25PGTDD271 pKa = 4.43DD272 pKa = 4.33IDD274 pKa = 5.47ADD276 pKa = 4.06SSPDD280 pKa = 4.2DD281 pKa = 3.89VDD283 pKa = 4.94ADD285 pKa = 4.22SDD287 pKa = 4.06TDD289 pKa = 5.84DD290 pKa = 4.13IDD292 pKa = 5.89ADD294 pKa = 3.73SSTNNDD300 pKa = 3.41GADD303 pKa = 3.36TNTNIVGANTRR314 pKa = 11.84TNNASADD321 pKa = 3.55PRR323 pKa = 11.84ANNAGADD330 pKa = 3.75SSANDD335 pKa = 3.54HH336 pKa = 6.96DD337 pKa = 5.33ANPNADD343 pKa = 3.89DD344 pKa = 4.48FGAGSSPDD352 pKa = 3.49DD353 pKa = 4.33HH354 pKa = 6.6GTDD357 pKa = 4.06PGTDD361 pKa = 4.32DD362 pKa = 4.31IDD364 pKa = 5.38ADD366 pKa = 3.76SSTDD370 pKa = 3.9FVSDD374 pKa = 3.63PTADD378 pKa = 3.56NTPEE382 pKa = 4.05PTPVPTTLVPTPLPTTLAPTPVPTTLAPTPAPTTLAPTPVPTTAAPAPTTGAPVNFDD439 pKa = 5.67LIDD442 pKa = 4.11CNEE445 pKa = 4.6NISIDD450 pKa = 3.54AMSAGIYY457 pKa = 10.12SVLDD461 pKa = 3.57KK462 pKa = 11.25TCMTEE467 pKa = 4.44FDD469 pKa = 4.7PLSEE473 pKa = 4.29TTTCMTKK480 pKa = 9.99YY481 pKa = 10.48CRR483 pKa = 11.84LCKK486 pKa = 10.44VADD489 pKa = 3.88TEE491 pKa = 4.43DD492 pKa = 3.79TVGLKK497 pKa = 10.1SCSQFFSFRR506 pKa = 11.84RR507 pKa = 11.84HH508 pKa = 4.51LVRR511 pKa = 11.84RR512 pKa = 11.84AAA514 pKa = 3.78
MM1 pKa = 7.4TKK3 pKa = 10.1SLHH6 pKa = 5.76LSXVAVTAAAVALLVDD22 pKa = 3.73SGIDD26 pKa = 3.05RR27 pKa = 11.84FGSVGVFAQTTGTAAATVAICRR49 pKa = 11.84APASSLTDD57 pKa = 3.3YY58 pKa = 11.35GVGIVSDD65 pKa = 4.48PACATPSSAASSSCIDD81 pKa = 3.63DD82 pKa = 3.14QCRR85 pKa = 11.84FCQVFRR91 pKa = 11.84TDD93 pKa = 3.2LSKK96 pKa = 11.21GYY98 pKa = 9.79LPCTTPPVLVSGGAGVPPSEE118 pKa = 4.57TPPSPSVPPTTLVPTLASALDD139 pKa = 3.87PTPTAPAAVTFEE151 pKa = 4.29PTVVAPAPASVSMTPVPSAVGIALGAVPATAPPISPLLIDD191 pKa = 4.63CSAEE195 pKa = 3.87LSLEE199 pKa = 3.9AMAAGVYY206 pKa = 10.15AVLDD210 pKa = 4.3PSCQADD216 pKa = 3.71DD217 pKa = 4.54DD218 pKa = 4.49AAQASCQDD226 pKa = 3.64EE227 pKa = 4.56FCRR230 pKa = 11.84LCRR233 pKa = 11.84VGEE236 pKa = 3.95SDD238 pKa = 3.97YY239 pKa = 11.81AANTDD244 pKa = 4.63DD245 pKa = 5.38AGADD249 pKa = 3.82PGTDD253 pKa = 3.58DD254 pKa = 4.57IGANSSADD262 pKa = 3.64NPGTDD267 pKa = 4.25PGTDD271 pKa = 4.43DD272 pKa = 4.33IDD274 pKa = 5.47ADD276 pKa = 4.06SSPDD280 pKa = 4.2DD281 pKa = 3.89VDD283 pKa = 4.94ADD285 pKa = 4.22SDD287 pKa = 4.06TDD289 pKa = 5.84DD290 pKa = 4.13IDD292 pKa = 5.89ADD294 pKa = 3.73SSTNNDD300 pKa = 3.41GADD303 pKa = 3.36TNTNIVGANTRR314 pKa = 11.84TNNASADD321 pKa = 3.55PRR323 pKa = 11.84ANNAGADD330 pKa = 3.75SSANDD335 pKa = 3.54HH336 pKa = 6.96DD337 pKa = 5.33ANPNADD343 pKa = 3.89DD344 pKa = 4.48FGAGSSPDD352 pKa = 3.49DD353 pKa = 4.33HH354 pKa = 6.6GTDD357 pKa = 4.06PGTDD361 pKa = 4.32DD362 pKa = 4.31IDD364 pKa = 5.38ADD366 pKa = 3.76SSTDD370 pKa = 3.9FVSDD374 pKa = 3.63PTADD378 pKa = 3.56NTPEE382 pKa = 4.05PTPVPTTLVPTPLPTTLAPTPVPTTLAPTPAPTTLAPTPVPTTAAPAPTTGAPVNFDD439 pKa = 5.67LIDD442 pKa = 4.11CNEE445 pKa = 4.6NISIDD450 pKa = 3.54AMSAGIYY457 pKa = 10.12SVLDD461 pKa = 3.57KK462 pKa = 11.25TCMTEE467 pKa = 4.44FDD469 pKa = 4.7PLSEE473 pKa = 4.29TTTCMTKK480 pKa = 9.99YY481 pKa = 10.48CRR483 pKa = 11.84LCKK486 pKa = 10.44VADD489 pKa = 3.88TEE491 pKa = 4.43DD492 pKa = 3.79TVGLKK497 pKa = 10.1SCSQFFSFRR506 pKa = 11.84RR507 pKa = 11.84HH508 pKa = 4.51LVRR511 pKa = 11.84RR512 pKa = 11.84AAA514 pKa = 3.78
Molecular weight: 51.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5D6Y9F7|A0A5D6Y9F7_9STRA Uncharacterized protein OS=Pythium brassicum OX=1485010 GN=PybrP1_007566 PE=4 SV=1
MM1 pKa = 7.44RR2 pKa = 11.84QRR4 pKa = 11.84LRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84PCRR11 pKa = 11.84PRR13 pKa = 11.84VRR15 pKa = 11.84QRR17 pKa = 11.84LCRR20 pKa = 11.84PLACRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84CRR29 pKa = 11.84HH30 pKa = 4.86PVCRR34 pKa = 11.84PVRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84LPRR42 pKa = 11.84PRR44 pKa = 11.84VRR46 pKa = 11.84RR47 pKa = 11.84PPARR51 pKa = 11.84RR52 pKa = 11.84SASSTATTRR61 pKa = 11.84TTAPGTGRR69 pKa = 11.84RR70 pKa = 11.84TRR72 pKa = 11.84ASATT76 pKa = 3.39
MM1 pKa = 7.44RR2 pKa = 11.84QRR4 pKa = 11.84LRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84PCRR11 pKa = 11.84PRR13 pKa = 11.84VRR15 pKa = 11.84QRR17 pKa = 11.84LCRR20 pKa = 11.84PLACRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84CRR29 pKa = 11.84HH30 pKa = 4.86PVCRR34 pKa = 11.84PVRR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84LPRR42 pKa = 11.84PRR44 pKa = 11.84VRR46 pKa = 11.84RR47 pKa = 11.84PPARR51 pKa = 11.84RR52 pKa = 11.84SASSTATTRR61 pKa = 11.84TTAPGTGRR69 pKa = 11.84RR70 pKa = 11.84TRR72 pKa = 11.84ASATT76 pKa = 3.39
Molecular weight: 9.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6730507 |
8 |
7627 |
517.8 |
56.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.36 ± 0.034 | 1.599 ± 0.01 |
5.759 ± 0.013 | 5.94 ± 0.022 |
3.656 ± 0.015 | 6.459 ± 0.019 |
2.569 ± 0.011 | 3.121 ± 0.013 |
4.089 ± 0.017 | 9.782 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.886 ± 0.009 | 2.643 ± 0.011 |
4.999 ± 0.019 | 3.965 ± 0.014 |
7.237 ± 0.022 | 8.021 ± 0.027 |
5.23 ± 0.016 | 7.385 ± 0.02 |
1.12 ± 0.006 | 2.165 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
