
Cassava virus C
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Miaviricetes; Ourlivirales; Botourmiaviridae; Ourmiavirus
Average proteome isoelectric point is 8.32
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
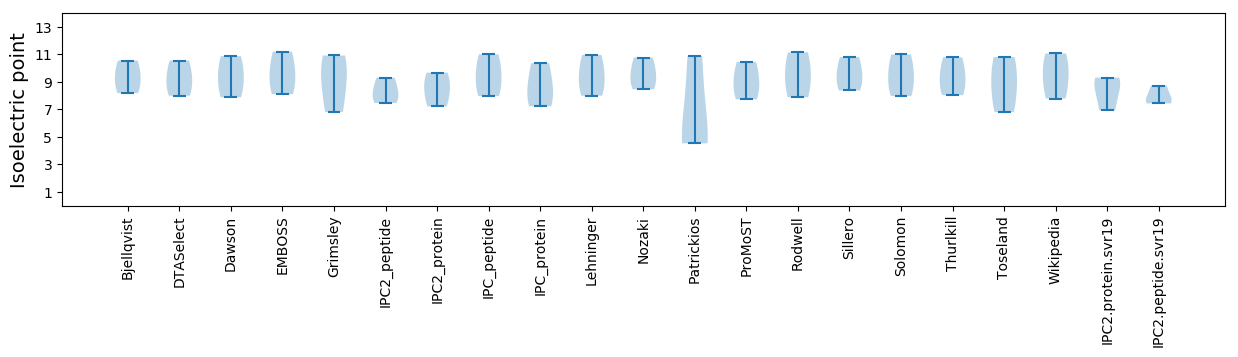
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|B5U1W8|B5U1W8_9VIRU Coat protein OS=Cassava virus C OX=561576 PE=2 SV=1
MM1 pKa = 7.78EE2 pKa = 5.48EE3 pKa = 5.04FPTIDD8 pKa = 3.51NRR10 pKa = 11.84IISADD15 pKa = 3.4LALPDD20 pKa = 4.27GNGNLAISIVEE31 pKa = 4.05PSSTLCLKK39 pKa = 10.76KK40 pKa = 10.18KK41 pKa = 10.13GHH43 pKa = 4.25VTYY46 pKa = 8.76RR47 pKa = 11.84TLMDD51 pKa = 4.79ASTSCQPQPLIPVSIWDD68 pKa = 3.76RR69 pKa = 11.84VRR71 pKa = 11.84LSAHH75 pKa = 5.49TAAHH79 pKa = 6.37RR80 pKa = 11.84NTYY83 pKa = 9.48VSYY86 pKa = 11.08EE87 pKa = 4.05EE88 pKa = 5.91VVAKK92 pKa = 8.95WVPSCLKK99 pKa = 10.74NEE101 pKa = 4.2AFGQFAVVDD110 pKa = 3.88TRR112 pKa = 11.84LQADD116 pKa = 4.14DD117 pKa = 4.9LEE119 pKa = 6.27DD120 pKa = 3.44MLRR123 pKa = 11.84RR124 pKa = 11.84AIWITDD130 pKa = 3.4EE131 pKa = 4.1VDD133 pKa = 3.12LGRR136 pKa = 11.84QYY138 pKa = 10.5TCSGALPFCLPVSSTDD154 pKa = 3.21RR155 pKa = 11.84GTINDD160 pKa = 3.67NPIKK164 pKa = 10.52VVLYY168 pKa = 6.54VTRR171 pKa = 11.84SGYY174 pKa = 9.93QEE176 pKa = 3.85AAKK179 pKa = 10.37LGSLTTTTKK188 pKa = 10.65LALSTSPIDD197 pKa = 4.31LVNTRR202 pKa = 11.84PAAFVEE208 pKa = 4.73KK209 pKa = 10.29KK210 pKa = 10.31AALKK214 pKa = 10.56IEE216 pKa = 4.13GTRR219 pKa = 11.84GGNLPSRR226 pKa = 11.84RR227 pKa = 11.84SSVEE231 pKa = 3.46TRR233 pKa = 11.84RR234 pKa = 11.84DD235 pKa = 3.26RR236 pKa = 11.84YY237 pKa = 10.36RR238 pKa = 11.84SGSARR243 pKa = 11.84WPARR247 pKa = 11.84DD248 pKa = 3.74RR249 pKa = 11.84DD250 pKa = 3.77EE251 pKa = 5.07SPTPGRR257 pKa = 11.84RR258 pKa = 11.84SFDD261 pKa = 3.44LPNRR265 pKa = 11.84LPVGGGTVPAAQDD278 pKa = 3.61PDD280 pKa = 4.2CFSSAGEE287 pKa = 4.11VGG289 pKa = 3.41
MM1 pKa = 7.78EE2 pKa = 5.48EE3 pKa = 5.04FPTIDD8 pKa = 3.51NRR10 pKa = 11.84IISADD15 pKa = 3.4LALPDD20 pKa = 4.27GNGNLAISIVEE31 pKa = 4.05PSSTLCLKK39 pKa = 10.76KK40 pKa = 10.18KK41 pKa = 10.13GHH43 pKa = 4.25VTYY46 pKa = 8.76RR47 pKa = 11.84TLMDD51 pKa = 4.79ASTSCQPQPLIPVSIWDD68 pKa = 3.76RR69 pKa = 11.84VRR71 pKa = 11.84LSAHH75 pKa = 5.49TAAHH79 pKa = 6.37RR80 pKa = 11.84NTYY83 pKa = 9.48VSYY86 pKa = 11.08EE87 pKa = 4.05EE88 pKa = 5.91VVAKK92 pKa = 8.95WVPSCLKK99 pKa = 10.74NEE101 pKa = 4.2AFGQFAVVDD110 pKa = 3.88TRR112 pKa = 11.84LQADD116 pKa = 4.14DD117 pKa = 4.9LEE119 pKa = 6.27DD120 pKa = 3.44MLRR123 pKa = 11.84RR124 pKa = 11.84AIWITDD130 pKa = 3.4EE131 pKa = 4.1VDD133 pKa = 3.12LGRR136 pKa = 11.84QYY138 pKa = 10.5TCSGALPFCLPVSSTDD154 pKa = 3.21RR155 pKa = 11.84GTINDD160 pKa = 3.67NPIKK164 pKa = 10.52VVLYY168 pKa = 6.54VTRR171 pKa = 11.84SGYY174 pKa = 9.93QEE176 pKa = 3.85AAKK179 pKa = 10.37LGSLTTTTKK188 pKa = 10.65LALSTSPIDD197 pKa = 4.31LVNTRR202 pKa = 11.84PAAFVEE208 pKa = 4.73KK209 pKa = 10.29KK210 pKa = 10.31AALKK214 pKa = 10.56IEE216 pKa = 4.13GTRR219 pKa = 11.84GGNLPSRR226 pKa = 11.84RR227 pKa = 11.84SSVEE231 pKa = 3.46TRR233 pKa = 11.84RR234 pKa = 11.84DD235 pKa = 3.26RR236 pKa = 11.84YY237 pKa = 10.36RR238 pKa = 11.84SGSARR243 pKa = 11.84WPARR247 pKa = 11.84DD248 pKa = 3.74RR249 pKa = 11.84DD250 pKa = 3.77EE251 pKa = 5.07SPTPGRR257 pKa = 11.84RR258 pKa = 11.84SFDD261 pKa = 3.44LPNRR265 pKa = 11.84LPVGGGTVPAAQDD278 pKa = 3.61PDD280 pKa = 4.2CFSSAGEE287 pKa = 4.11VGG289 pKa = 3.41
Molecular weight: 31.6 kDa
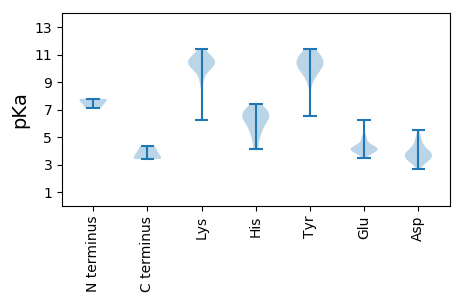
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5U1W8|B5U1W8_9VIRU Coat protein OS=Cassava virus C OX=561576 PE=2 SV=1
MM1 pKa = 7.11VARR4 pKa = 11.84SNRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84QNRR13 pKa = 11.84QPQGQITRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84NGGGNNNNPPRR34 pKa = 11.84SIPRR38 pKa = 11.84LAGTVIKK45 pKa = 10.41EE46 pKa = 4.46SIFEE50 pKa = 4.19SSFDD54 pKa = 3.97LDD56 pKa = 3.86LDD58 pKa = 3.76KK59 pKa = 11.34GANKK63 pKa = 10.47LKK65 pKa = 10.5VQALAFGPGATIGWTKK81 pKa = 10.1PPRR84 pKa = 11.84TIAWKK89 pKa = 10.31LKK91 pKa = 8.88TMQLDD96 pKa = 3.34WVSFLRR102 pKa = 11.84PTKK105 pKa = 10.34HH106 pKa = 6.54AGEE109 pKa = 4.32LKK111 pKa = 9.73ICLVPGRR118 pKa = 11.84GLNGLSKK125 pKa = 10.92DD126 pKa = 3.46QTLALAPAEE135 pKa = 4.13IARR138 pKa = 11.84SNSVRR143 pKa = 11.84TIRR146 pKa = 11.84VGGTLSRR153 pKa = 11.84NPWSGYY159 pKa = 6.71SQWVEE164 pKa = 3.78PPKK167 pKa = 10.69GVSNSGEE174 pKa = 4.28GEE176 pKa = 3.91NVAYY180 pKa = 9.83RR181 pKa = 11.84VVFDD185 pKa = 4.21PSNVLGTDD193 pKa = 3.04EE194 pKa = 4.59GEE196 pKa = 4.1KK197 pKa = 10.63LLRR200 pKa = 11.84VYY202 pKa = 10.44FKK204 pKa = 10.85LVYY207 pKa = 9.7QVQEE211 pKa = 3.95VAII214 pKa = 4.31
MM1 pKa = 7.11VARR4 pKa = 11.84SNRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84QNRR13 pKa = 11.84QPQGQITRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84NGGGNNNNPPRR34 pKa = 11.84SIPRR38 pKa = 11.84LAGTVIKK45 pKa = 10.41EE46 pKa = 4.46SIFEE50 pKa = 4.19SSFDD54 pKa = 3.97LDD56 pKa = 3.86LDD58 pKa = 3.76KK59 pKa = 11.34GANKK63 pKa = 10.47LKK65 pKa = 10.5VQALAFGPGATIGWTKK81 pKa = 10.1PPRR84 pKa = 11.84TIAWKK89 pKa = 10.31LKK91 pKa = 8.88TMQLDD96 pKa = 3.34WVSFLRR102 pKa = 11.84PTKK105 pKa = 10.34HH106 pKa = 6.54AGEE109 pKa = 4.32LKK111 pKa = 9.73ICLVPGRR118 pKa = 11.84GLNGLSKK125 pKa = 10.92DD126 pKa = 3.46QTLALAPAEE135 pKa = 4.13IARR138 pKa = 11.84SNSVRR143 pKa = 11.84TIRR146 pKa = 11.84VGGTLSRR153 pKa = 11.84NPWSGYY159 pKa = 6.71SQWVEE164 pKa = 3.78PPKK167 pKa = 10.69GVSNSGEE174 pKa = 4.28GEE176 pKa = 3.91NVAYY180 pKa = 9.83RR181 pKa = 11.84VVFDD185 pKa = 4.21PSNVLGTDD193 pKa = 3.04EE194 pKa = 4.59GEE196 pKa = 4.1KK197 pKa = 10.63LLRR200 pKa = 11.84VYY202 pKa = 10.44FKK204 pKa = 10.85LVYY207 pKa = 9.7QVQEE211 pKa = 3.95VAII214 pKa = 4.31
Molecular weight: 23.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1373 |
214 |
870 |
457.7 |
51.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.992 ± 0.577 | 1.748 ± 0.34 |
5.098 ± 0.689 | 5.39 ± 0.33 |
4.079 ± 0.806 | 7.065 ± 0.734 |
1.675 ± 0.475 | 5.171 ± 0.323 |
4.516 ± 0.428 | 9.905 ± 0.623 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.311 ± 0.169 | 3.714 ± 0.772 |
6.555 ± 0.133 | 3.132 ± 0.424 |
8.958 ± 0.127 | 8.012 ± 0.374 |
5.317 ± 0.952 | 6.919 ± 0.354 |
1.894 ± 0.186 | 2.549 ± 0.213 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
