
endosymbiont of Escarpia spicata
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
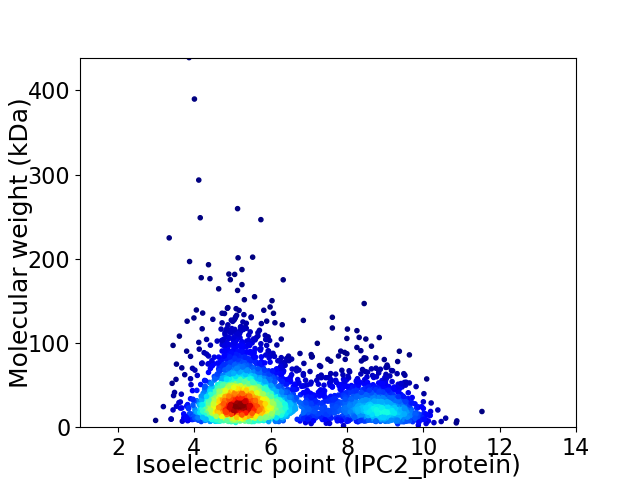
Virtual 2D-PAGE plot for 3499 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A370DQB1|A0A370DQB1_9GAMM TPR_REGION domain-containing protein OS=endosymbiont of Escarpia spicata OX=2200908 GN=DIZ78_05625 PE=4 SV=1
MM1 pKa = 7.34LVWQHH6 pKa = 6.57AEE8 pKa = 3.98SEE10 pKa = 4.33DD11 pKa = 4.11GISCVGISRR20 pKa = 11.84EE21 pKa = 4.12TGEE24 pKa = 5.37DD25 pKa = 3.22PMKK28 pKa = 10.94AIILALALVGVLFAPSTALAEE49 pKa = 4.13DD50 pKa = 4.02TPLDD54 pKa = 3.52KK55 pKa = 10.84TVWTTSEE62 pKa = 4.33STWWWSLWPEE72 pKa = 4.03PVSCDD77 pKa = 2.84GQAFHH82 pKa = 7.35LIDD85 pKa = 3.65GVYY88 pKa = 9.28TYY90 pKa = 10.66SYY92 pKa = 10.03PADD95 pKa = 3.51PCGRR99 pKa = 11.84GRR101 pKa = 11.84SFFSPANALDD111 pKa = 3.74NHH113 pKa = 5.84GQWFQIDD120 pKa = 3.97FGVSLTLTRR129 pKa = 11.84IRR131 pKa = 11.84FYY133 pKa = 11.18QFVEE137 pKa = 4.11NTLMSSLPGTDD148 pKa = 2.51KK149 pKa = 10.62YY150 pKa = 11.55SRR152 pKa = 11.84YY153 pKa = 8.45INEE156 pKa = 3.71MRR158 pKa = 11.84FTFSDD163 pKa = 3.53GTEE166 pKa = 3.96EE167 pKa = 4.99LVAFPPGTPFSTRR180 pKa = 11.84EE181 pKa = 3.87SVTYY185 pKa = 10.95ADD187 pKa = 4.64FAAPHH192 pKa = 4.77TTEE195 pKa = 4.72SIRR198 pKa = 11.84ATIVSFHH205 pKa = 6.71NDD207 pKa = 2.75YY208 pKa = 11.51SLAGLYY214 pKa = 10.48ALEE217 pKa = 4.76IDD219 pKa = 4.44FFGAAGDD226 pKa = 4.34PDD228 pKa = 5.29DD229 pKa = 5.96LDD231 pKa = 5.58GDD233 pKa = 4.69GIVNADD239 pKa = 4.1DD240 pKa = 4.14NCPLSANPDD249 pKa = 3.85QLDD252 pKa = 3.56MDD254 pKa = 4.67HH255 pKa = 7.4DD256 pKa = 5.51GIGNICDD263 pKa = 4.06DD264 pKa = 5.67DD265 pKa = 4.65IDD267 pKa = 4.75GDD269 pKa = 3.89EE270 pKa = 4.65FANVDD275 pKa = 4.15DD276 pKa = 4.93NCPEE280 pKa = 4.17AANSLQSDD288 pKa = 3.91GDD290 pKa = 3.78LDD292 pKa = 5.11GIGDD296 pKa = 4.06ACDD299 pKa = 3.87PEE301 pKa = 4.85CNPRR305 pKa = 11.84LTVKK309 pKa = 10.7DD310 pKa = 3.53GWIGNQSSWWLQPWRR325 pKa = 11.84NTCDD329 pKa = 3.24GKK331 pKa = 10.97AQNAIDD337 pKa = 3.5GNYY340 pKa = 9.49RR341 pKa = 11.84FTYY344 pKa = 9.38SNYY347 pKa = 8.11WACNGDD353 pKa = 3.84LSPRR357 pKa = 11.84SFFAPRR363 pKa = 11.84FTGTDD368 pKa = 2.81VGTWMQVEE376 pKa = 4.68FPEE379 pKa = 4.7VYY381 pKa = 9.51TINGITLAQIKK392 pKa = 9.67EE393 pKa = 3.94RR394 pKa = 11.84TYY396 pKa = 10.5VSGLVRR402 pKa = 11.84PHH404 pKa = 7.29KK405 pKa = 10.61YY406 pKa = 9.89GAYY409 pKa = 10.22LKK411 pKa = 10.61DD412 pKa = 3.41AMVEE416 pKa = 4.13FSDD419 pKa = 4.29GSSVEE424 pKa = 3.98VTFPQQLEE432 pKa = 4.03ASVTFPEE439 pKa = 4.44IATQSVHH446 pKa = 3.36ITARR450 pKa = 11.84SFHH453 pKa = 6.2PTVTTNPAWLVVEE466 pKa = 4.78ADD468 pKa = 3.66FAGAPGDD475 pKa = 3.3VDD477 pKa = 5.64LNLEE481 pKa = 4.37VCSEE485 pKa = 4.16PAPLPPDD492 pKa = 3.46TDD494 pKa = 3.8GDD496 pKa = 4.35GVPDD500 pKa = 4.48HH501 pKa = 7.32LDD503 pKa = 3.52AFPNDD508 pKa = 3.46PTEE511 pKa = 3.97WSDD514 pKa = 3.74VDD516 pKa = 3.8GDD518 pKa = 4.68GIGDD522 pKa = 3.79NADD525 pKa = 4.0PDD527 pKa = 4.52DD528 pKa = 5.83DD529 pKa = 5.09NDD531 pKa = 5.78GIPDD535 pKa = 4.72DD536 pKa = 5.0EE537 pKa = 4.66DD538 pKa = 4.22TNPTEE543 pKa = 4.8ADD545 pKa = 3.36ALEE548 pKa = 5.2DD549 pKa = 3.75SDD551 pKa = 5.98GDD553 pKa = 4.3GIPNQDD559 pKa = 5.65DD560 pKa = 4.07PDD562 pKa = 5.41DD563 pKa = 5.9DD564 pKa = 4.48NDD566 pKa = 4.04GVLDD570 pKa = 4.18EE571 pKa = 6.33DD572 pKa = 4.91DD573 pKa = 5.57AFSKK577 pKa = 11.1DD578 pKa = 3.3PTEE581 pKa = 4.14WADD584 pKa = 3.3TDD586 pKa = 3.88GDD588 pKa = 4.48GVGDD592 pKa = 4.13NSDD595 pKa = 3.53NCITGYY601 pKa = 10.95NPGQADD607 pKa = 3.94ADD609 pKa = 3.75NDD611 pKa = 5.09GIGDD615 pKa = 3.78PCDD618 pKa = 3.85RR619 pKa = 11.84PIANAGMDD627 pKa = 3.65QSAHH631 pKa = 6.86PGTAVTLDD639 pKa = 3.78GTGSTDD645 pKa = 3.34PDD647 pKa = 3.31EE648 pKa = 5.14EE649 pKa = 4.6YY650 pKa = 10.43PLLFAWQLISAPPGSLASVQNPTSATPSLTPDD682 pKa = 2.89MMGDD686 pKa = 3.63YY687 pKa = 10.66LLEE690 pKa = 4.33LSVTDD695 pKa = 3.79QQGHH699 pKa = 5.26VSDD702 pKa = 4.12SDD704 pKa = 3.34KK705 pKa = 11.73VMVSTVNTAPTADD718 pKa = 3.75AGEE721 pKa = 4.68DD722 pKa = 3.47LAITQTGQWVALSGLASYY740 pKa = 11.15DD741 pKa = 3.65LDD743 pKa = 5.86GDD745 pKa = 4.99DD746 pKa = 5.63LIAYY750 pKa = 9.0QWMLVSKK757 pKa = 10.14PDD759 pKa = 3.38GSLAEE764 pKa = 4.31PAPADD769 pKa = 3.61TVEE772 pKa = 4.15TLFLADD778 pKa = 4.82LYY780 pKa = 11.12GSYY783 pKa = 10.7VFEE786 pKa = 4.34LTVTDD791 pKa = 4.28AYY793 pKa = 11.38YY794 pKa = 10.78SVSEE798 pKa = 4.12PDD800 pKa = 3.42QITVSLANVRR810 pKa = 11.84PVADD814 pKa = 4.18AGDD817 pKa = 3.82NLSVYY822 pKa = 10.27VGEE825 pKa = 4.42SVSLGGTGSHH835 pKa = 7.52DD836 pKa = 4.04ANGDD840 pKa = 3.53PLTYY844 pKa = 9.9HH845 pKa = 7.02WSLVLKK851 pKa = 10.17PDD853 pKa = 3.32GSTAEE858 pKa = 4.36LAAPDD863 pKa = 3.55QAMTGFMADD872 pKa = 2.99LAGTYY877 pKa = 9.79LVSLVVNDD885 pKa = 4.44GLQDD889 pKa = 3.82SEE891 pKa = 4.49PSSVTVTALTRR902 pKa = 11.84QDD904 pKa = 3.49EE905 pKa = 4.76VVTLVQDD912 pKa = 3.37VMDD915 pKa = 5.99RR916 pKa = 11.84INGFDD921 pKa = 3.25SSVFKK926 pKa = 11.05NKK928 pKa = 9.79NLAKK932 pKa = 10.55PFTNKK937 pKa = 9.72LNAALSKK944 pKa = 10.73LDD946 pKa = 3.63NGDD949 pKa = 3.46YY950 pKa = 11.35ADD952 pKa = 4.64ALNKK956 pKa = 10.2LEE958 pKa = 5.28HH959 pKa = 7.47DD960 pKa = 4.29ILPKK964 pKa = 9.84IDD966 pKa = 3.42GCATSGQPDD975 pKa = 3.58KK976 pKa = 11.25QDD978 pKa = 2.74WLTTCEE984 pKa = 4.17AQAEE988 pKa = 4.66VYY990 pKa = 10.42PLVQQAIEE998 pKa = 4.21LLRR1001 pKa = 11.84SLII1004 pKa = 4.0
MM1 pKa = 7.34LVWQHH6 pKa = 6.57AEE8 pKa = 3.98SEE10 pKa = 4.33DD11 pKa = 4.11GISCVGISRR20 pKa = 11.84EE21 pKa = 4.12TGEE24 pKa = 5.37DD25 pKa = 3.22PMKK28 pKa = 10.94AIILALALVGVLFAPSTALAEE49 pKa = 4.13DD50 pKa = 4.02TPLDD54 pKa = 3.52KK55 pKa = 10.84TVWTTSEE62 pKa = 4.33STWWWSLWPEE72 pKa = 4.03PVSCDD77 pKa = 2.84GQAFHH82 pKa = 7.35LIDD85 pKa = 3.65GVYY88 pKa = 9.28TYY90 pKa = 10.66SYY92 pKa = 10.03PADD95 pKa = 3.51PCGRR99 pKa = 11.84GRR101 pKa = 11.84SFFSPANALDD111 pKa = 3.74NHH113 pKa = 5.84GQWFQIDD120 pKa = 3.97FGVSLTLTRR129 pKa = 11.84IRR131 pKa = 11.84FYY133 pKa = 11.18QFVEE137 pKa = 4.11NTLMSSLPGTDD148 pKa = 2.51KK149 pKa = 10.62YY150 pKa = 11.55SRR152 pKa = 11.84YY153 pKa = 8.45INEE156 pKa = 3.71MRR158 pKa = 11.84FTFSDD163 pKa = 3.53GTEE166 pKa = 3.96EE167 pKa = 4.99LVAFPPGTPFSTRR180 pKa = 11.84EE181 pKa = 3.87SVTYY185 pKa = 10.95ADD187 pKa = 4.64FAAPHH192 pKa = 4.77TTEE195 pKa = 4.72SIRR198 pKa = 11.84ATIVSFHH205 pKa = 6.71NDD207 pKa = 2.75YY208 pKa = 11.51SLAGLYY214 pKa = 10.48ALEE217 pKa = 4.76IDD219 pKa = 4.44FFGAAGDD226 pKa = 4.34PDD228 pKa = 5.29DD229 pKa = 5.96LDD231 pKa = 5.58GDD233 pKa = 4.69GIVNADD239 pKa = 4.1DD240 pKa = 4.14NCPLSANPDD249 pKa = 3.85QLDD252 pKa = 3.56MDD254 pKa = 4.67HH255 pKa = 7.4DD256 pKa = 5.51GIGNICDD263 pKa = 4.06DD264 pKa = 5.67DD265 pKa = 4.65IDD267 pKa = 4.75GDD269 pKa = 3.89EE270 pKa = 4.65FANVDD275 pKa = 4.15DD276 pKa = 4.93NCPEE280 pKa = 4.17AANSLQSDD288 pKa = 3.91GDD290 pKa = 3.78LDD292 pKa = 5.11GIGDD296 pKa = 4.06ACDD299 pKa = 3.87PEE301 pKa = 4.85CNPRR305 pKa = 11.84LTVKK309 pKa = 10.7DD310 pKa = 3.53GWIGNQSSWWLQPWRR325 pKa = 11.84NTCDD329 pKa = 3.24GKK331 pKa = 10.97AQNAIDD337 pKa = 3.5GNYY340 pKa = 9.49RR341 pKa = 11.84FTYY344 pKa = 9.38SNYY347 pKa = 8.11WACNGDD353 pKa = 3.84LSPRR357 pKa = 11.84SFFAPRR363 pKa = 11.84FTGTDD368 pKa = 2.81VGTWMQVEE376 pKa = 4.68FPEE379 pKa = 4.7VYY381 pKa = 9.51TINGITLAQIKK392 pKa = 9.67EE393 pKa = 3.94RR394 pKa = 11.84TYY396 pKa = 10.5VSGLVRR402 pKa = 11.84PHH404 pKa = 7.29KK405 pKa = 10.61YY406 pKa = 9.89GAYY409 pKa = 10.22LKK411 pKa = 10.61DD412 pKa = 3.41AMVEE416 pKa = 4.13FSDD419 pKa = 4.29GSSVEE424 pKa = 3.98VTFPQQLEE432 pKa = 4.03ASVTFPEE439 pKa = 4.44IATQSVHH446 pKa = 3.36ITARR450 pKa = 11.84SFHH453 pKa = 6.2PTVTTNPAWLVVEE466 pKa = 4.78ADD468 pKa = 3.66FAGAPGDD475 pKa = 3.3VDD477 pKa = 5.64LNLEE481 pKa = 4.37VCSEE485 pKa = 4.16PAPLPPDD492 pKa = 3.46TDD494 pKa = 3.8GDD496 pKa = 4.35GVPDD500 pKa = 4.48HH501 pKa = 7.32LDD503 pKa = 3.52AFPNDD508 pKa = 3.46PTEE511 pKa = 3.97WSDD514 pKa = 3.74VDD516 pKa = 3.8GDD518 pKa = 4.68GIGDD522 pKa = 3.79NADD525 pKa = 4.0PDD527 pKa = 4.52DD528 pKa = 5.83DD529 pKa = 5.09NDD531 pKa = 5.78GIPDD535 pKa = 4.72DD536 pKa = 5.0EE537 pKa = 4.66DD538 pKa = 4.22TNPTEE543 pKa = 4.8ADD545 pKa = 3.36ALEE548 pKa = 5.2DD549 pKa = 3.75SDD551 pKa = 5.98GDD553 pKa = 4.3GIPNQDD559 pKa = 5.65DD560 pKa = 4.07PDD562 pKa = 5.41DD563 pKa = 5.9DD564 pKa = 4.48NDD566 pKa = 4.04GVLDD570 pKa = 4.18EE571 pKa = 6.33DD572 pKa = 4.91DD573 pKa = 5.57AFSKK577 pKa = 11.1DD578 pKa = 3.3PTEE581 pKa = 4.14WADD584 pKa = 3.3TDD586 pKa = 3.88GDD588 pKa = 4.48GVGDD592 pKa = 4.13NSDD595 pKa = 3.53NCITGYY601 pKa = 10.95NPGQADD607 pKa = 3.94ADD609 pKa = 3.75NDD611 pKa = 5.09GIGDD615 pKa = 3.78PCDD618 pKa = 3.85RR619 pKa = 11.84PIANAGMDD627 pKa = 3.65QSAHH631 pKa = 6.86PGTAVTLDD639 pKa = 3.78GTGSTDD645 pKa = 3.34PDD647 pKa = 3.31EE648 pKa = 5.14EE649 pKa = 4.6YY650 pKa = 10.43PLLFAWQLISAPPGSLASVQNPTSATPSLTPDD682 pKa = 2.89MMGDD686 pKa = 3.63YY687 pKa = 10.66LLEE690 pKa = 4.33LSVTDD695 pKa = 3.79QQGHH699 pKa = 5.26VSDD702 pKa = 4.12SDD704 pKa = 3.34KK705 pKa = 11.73VMVSTVNTAPTADD718 pKa = 3.75AGEE721 pKa = 4.68DD722 pKa = 3.47LAITQTGQWVALSGLASYY740 pKa = 11.15DD741 pKa = 3.65LDD743 pKa = 5.86GDD745 pKa = 4.99DD746 pKa = 5.63LIAYY750 pKa = 9.0QWMLVSKK757 pKa = 10.14PDD759 pKa = 3.38GSLAEE764 pKa = 4.31PAPADD769 pKa = 3.61TVEE772 pKa = 4.15TLFLADD778 pKa = 4.82LYY780 pKa = 11.12GSYY783 pKa = 10.7VFEE786 pKa = 4.34LTVTDD791 pKa = 4.28AYY793 pKa = 11.38YY794 pKa = 10.78SVSEE798 pKa = 4.12PDD800 pKa = 3.42QITVSLANVRR810 pKa = 11.84PVADD814 pKa = 4.18AGDD817 pKa = 3.82NLSVYY822 pKa = 10.27VGEE825 pKa = 4.42SVSLGGTGSHH835 pKa = 7.52DD836 pKa = 4.04ANGDD840 pKa = 3.53PLTYY844 pKa = 9.9HH845 pKa = 7.02WSLVLKK851 pKa = 10.17PDD853 pKa = 3.32GSTAEE858 pKa = 4.36LAAPDD863 pKa = 3.55QAMTGFMADD872 pKa = 2.99LAGTYY877 pKa = 9.79LVSLVVNDD885 pKa = 4.44GLQDD889 pKa = 3.82SEE891 pKa = 4.49PSSVTVTALTRR902 pKa = 11.84QDD904 pKa = 3.49EE905 pKa = 4.76VVTLVQDD912 pKa = 3.37VMDD915 pKa = 5.99RR916 pKa = 11.84INGFDD921 pKa = 3.25SSVFKK926 pKa = 11.05NKK928 pKa = 9.79NLAKK932 pKa = 10.55PFTNKK937 pKa = 9.72LNAALSKK944 pKa = 10.73LDD946 pKa = 3.63NGDD949 pKa = 3.46YY950 pKa = 11.35ADD952 pKa = 4.64ALNKK956 pKa = 10.2LEE958 pKa = 5.28HH959 pKa = 7.47DD960 pKa = 4.29ILPKK964 pKa = 9.84IDD966 pKa = 3.42GCATSGQPDD975 pKa = 3.58KK976 pKa = 11.25QDD978 pKa = 2.74WLTTCEE984 pKa = 4.17AQAEE988 pKa = 4.66VYY990 pKa = 10.42PLVQQAIEE998 pKa = 4.21LLRR1001 pKa = 11.84SLII1004 pKa = 4.0
Molecular weight: 108.13 kDa
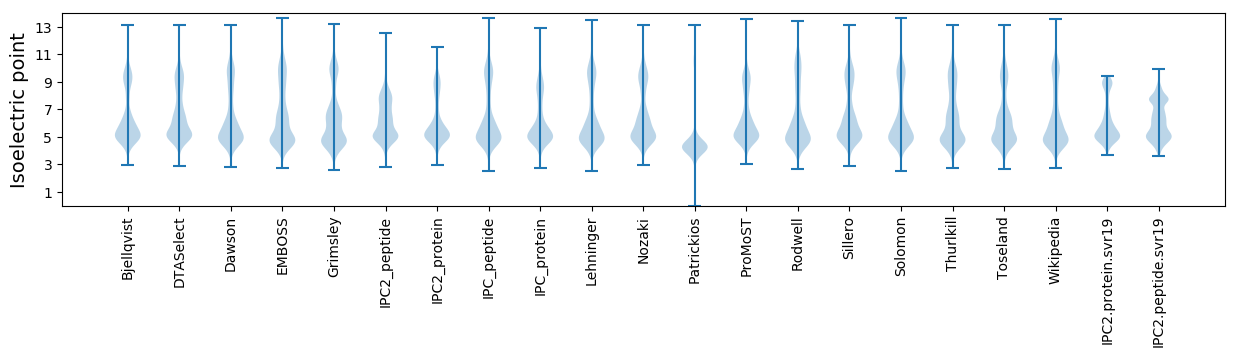
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A370DR43|A0A370DR43_9GAMM YceI family protein OS=endosymbiont of Escarpia spicata OX=2200908 GN=DIZ78_06040 PE=4 SV=1
MM1 pKa = 7.6AKK3 pKa = 10.21KK4 pKa = 8.94KK5 pKa = 7.63TAKK8 pKa = 10.23KK9 pKa = 8.3KK10 pKa = 7.97TAVKK14 pKa = 10.21RR15 pKa = 11.84KK16 pKa = 8.79VSTTKK21 pKa = 10.37RR22 pKa = 11.84VAAKK26 pKa = 10.13KK27 pKa = 9.91KK28 pKa = 9.71ASTRR32 pKa = 11.84KK33 pKa = 9.74KK34 pKa = 10.4SVAKK38 pKa = 10.45KK39 pKa = 9.92SASKK43 pKa = 10.51KK44 pKa = 9.65KK45 pKa = 10.54AVTKK49 pKa = 10.55KK50 pKa = 9.21KK51 pKa = 8.18TVKK54 pKa = 10.09KK55 pKa = 9.86RR56 pKa = 11.84AAKK59 pKa = 10.18KK60 pKa = 10.03KK61 pKa = 9.24VAAKK65 pKa = 10.13KK66 pKa = 8.99KK67 pKa = 7.06TVKK70 pKa = 10.12KK71 pKa = 9.95RR72 pKa = 11.84AAKK75 pKa = 10.18KK76 pKa = 10.03KK77 pKa = 9.24VAAKK81 pKa = 10.13KK82 pKa = 8.99KK83 pKa = 7.06TVKK86 pKa = 10.12KK87 pKa = 9.95RR88 pKa = 11.84AAKK91 pKa = 10.18KK92 pKa = 10.03KK93 pKa = 9.24VAAKK97 pKa = 10.13KK98 pKa = 8.99KK99 pKa = 7.06TVKK102 pKa = 10.12KK103 pKa = 9.95RR104 pKa = 11.84AAKK107 pKa = 10.18KK108 pKa = 10.03KK109 pKa = 9.24VAAKK113 pKa = 10.13KK114 pKa = 8.99KK115 pKa = 7.06TVKK118 pKa = 10.12KK119 pKa = 9.95RR120 pKa = 11.84AAKK123 pKa = 10.18KK124 pKa = 10.03KK125 pKa = 9.24VAAKK129 pKa = 10.13KK130 pKa = 8.99KK131 pKa = 7.06TVKK134 pKa = 10.12KK135 pKa = 9.95RR136 pKa = 11.84AAKK139 pKa = 10.18KK140 pKa = 10.03KK141 pKa = 9.24VAAKK145 pKa = 10.13KK146 pKa = 8.99KK147 pKa = 7.06TVKK150 pKa = 10.08KK151 pKa = 9.89RR152 pKa = 11.84AARR155 pKa = 11.84PRR157 pKa = 11.84KK158 pKa = 7.53VTPRR162 pKa = 11.84KK163 pKa = 9.84KK164 pKa = 9.64ATQKK168 pKa = 11.07
MM1 pKa = 7.6AKK3 pKa = 10.21KK4 pKa = 8.94KK5 pKa = 7.63TAKK8 pKa = 10.23KK9 pKa = 8.3KK10 pKa = 7.97TAVKK14 pKa = 10.21RR15 pKa = 11.84KK16 pKa = 8.79VSTTKK21 pKa = 10.37RR22 pKa = 11.84VAAKK26 pKa = 10.13KK27 pKa = 9.91KK28 pKa = 9.71ASTRR32 pKa = 11.84KK33 pKa = 9.74KK34 pKa = 10.4SVAKK38 pKa = 10.45KK39 pKa = 9.92SASKK43 pKa = 10.51KK44 pKa = 9.65KK45 pKa = 10.54AVTKK49 pKa = 10.55KK50 pKa = 9.21KK51 pKa = 8.18TVKK54 pKa = 10.09KK55 pKa = 9.86RR56 pKa = 11.84AAKK59 pKa = 10.18KK60 pKa = 10.03KK61 pKa = 9.24VAAKK65 pKa = 10.13KK66 pKa = 8.99KK67 pKa = 7.06TVKK70 pKa = 10.12KK71 pKa = 9.95RR72 pKa = 11.84AAKK75 pKa = 10.18KK76 pKa = 10.03KK77 pKa = 9.24VAAKK81 pKa = 10.13KK82 pKa = 8.99KK83 pKa = 7.06TVKK86 pKa = 10.12KK87 pKa = 9.95RR88 pKa = 11.84AAKK91 pKa = 10.18KK92 pKa = 10.03KK93 pKa = 9.24VAAKK97 pKa = 10.13KK98 pKa = 8.99KK99 pKa = 7.06TVKK102 pKa = 10.12KK103 pKa = 9.95RR104 pKa = 11.84AAKK107 pKa = 10.18KK108 pKa = 10.03KK109 pKa = 9.24VAAKK113 pKa = 10.13KK114 pKa = 8.99KK115 pKa = 7.06TVKK118 pKa = 10.12KK119 pKa = 9.95RR120 pKa = 11.84AAKK123 pKa = 10.18KK124 pKa = 10.03KK125 pKa = 9.24VAAKK129 pKa = 10.13KK130 pKa = 8.99KK131 pKa = 7.06TVKK134 pKa = 10.12KK135 pKa = 9.95RR136 pKa = 11.84AAKK139 pKa = 10.18KK140 pKa = 10.03KK141 pKa = 9.24VAAKK145 pKa = 10.13KK146 pKa = 8.99KK147 pKa = 7.06TVKK150 pKa = 10.08KK151 pKa = 9.89RR152 pKa = 11.84AARR155 pKa = 11.84PRR157 pKa = 11.84KK158 pKa = 7.53VTPRR162 pKa = 11.84KK163 pKa = 9.84KK164 pKa = 9.64ATQKK168 pKa = 11.07
Molecular weight: 18.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1169978 |
17 |
4119 |
334.4 |
37.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.017 ± 0.04 | 0.979 ± 0.017 |
5.918 ± 0.035 | 6.687 ± 0.042 |
3.783 ± 0.028 | 7.694 ± 0.044 |
2.264 ± 0.022 | 5.746 ± 0.035 |
4.284 ± 0.041 | 10.816 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.522 ± 0.024 | 3.448 ± 0.029 |
4.537 ± 0.03 | 4.098 ± 0.034 |
6.164 ± 0.047 | 6.124 ± 0.032 |
5.21 ± 0.044 | 6.787 ± 0.04 |
1.285 ± 0.016 | 2.635 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
