
Infectious salmon anemia virus (isolate Atlantic salmon/Norway/810/9/99) (ISAV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Isavirus; Salmon isavirus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
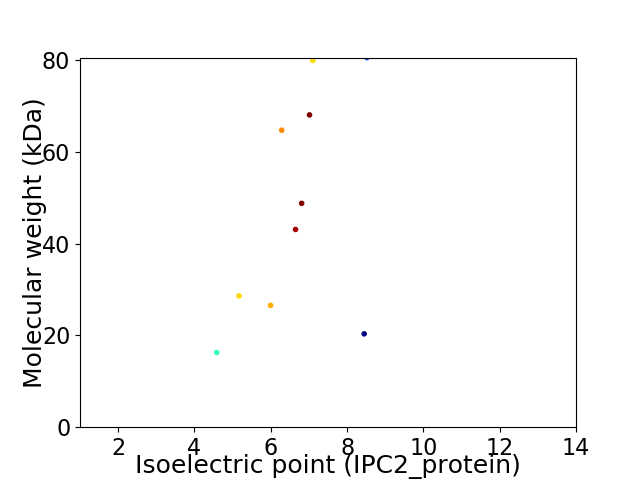
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


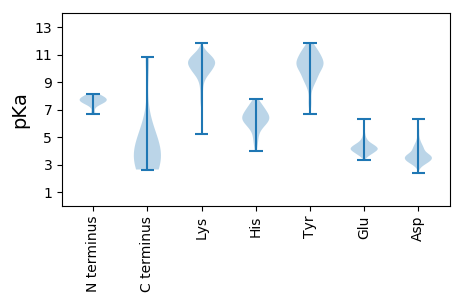
Protein with the lowest isoelectric point:
>sp|Q8V3U2|NS1_ISAV8 Non-structural protein 1 OS=Infectious salmon anemia virus (isolate Atlantic salmon/Norway/810/9/99) OX=652965 GN=Segment-7 PE=4 SV=1
MM1 pKa = 7.44NLLLLLQVASFLSDD15 pKa = 3.19SKK17 pKa = 11.8VPGEE21 pKa = 5.17DD22 pKa = 3.39GTSSTSGMLDD32 pKa = 3.61LLRR35 pKa = 11.84DD36 pKa = 3.77QVDD39 pKa = 3.62SLSINDD45 pKa = 3.7STTEE49 pKa = 3.78PKK51 pKa = 9.8TRR53 pKa = 11.84LDD55 pKa = 3.52PGLYY59 pKa = 9.38PWLKK63 pKa = 7.7WTEE66 pKa = 3.69TAYY69 pKa = 10.61RR70 pKa = 11.84SSTRR74 pKa = 11.84SLASTIVMGALVQQRR89 pKa = 11.84GSGNGITMRR98 pKa = 11.84EE99 pKa = 4.14LEE101 pKa = 4.67LSLGLDD107 pKa = 3.83FTSEE111 pKa = 4.25CDD113 pKa = 3.26WLKK116 pKa = 9.58TCYY119 pKa = 10.13VNKK122 pKa = 10.35NFVFLSEE129 pKa = 4.45KK130 pKa = 10.21EE131 pKa = 3.84IAVNMEE137 pKa = 3.8VEE139 pKa = 4.59KK140 pKa = 10.32FICNEE145 pKa = 3.63NN146 pKa = 3.3
MM1 pKa = 7.44NLLLLLQVASFLSDD15 pKa = 3.19SKK17 pKa = 11.8VPGEE21 pKa = 5.17DD22 pKa = 3.39GTSSTSGMLDD32 pKa = 3.61LLRR35 pKa = 11.84DD36 pKa = 3.77QVDD39 pKa = 3.62SLSINDD45 pKa = 3.7STTEE49 pKa = 3.78PKK51 pKa = 9.8TRR53 pKa = 11.84LDD55 pKa = 3.52PGLYY59 pKa = 9.38PWLKK63 pKa = 7.7WTEE66 pKa = 3.69TAYY69 pKa = 10.61RR70 pKa = 11.84SSTRR74 pKa = 11.84SLASTIVMGALVQQRR89 pKa = 11.84GSGNGITMRR98 pKa = 11.84EE99 pKa = 4.14LEE101 pKa = 4.67LSLGLDD107 pKa = 3.83FTSEE111 pKa = 4.25CDD113 pKa = 3.26WLKK116 pKa = 9.58TCYY119 pKa = 10.13VNKK122 pKa = 10.35NFVFLSEE129 pKa = 4.45KK130 pKa = 10.21EE131 pKa = 3.84IAVNMEE137 pKa = 3.8VEE139 pKa = 4.59KK140 pKa = 10.32FICNEE145 pKa = 3.63NN146 pKa = 3.3
Molecular weight: 16.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8V3T7|NCAP_ISAV8 Nucleoprotein OS=Infectious salmon anemia virus (isolate Atlantic salmon/Norway/810/9/99) OX=652965 GN=Segment-3 PE=1 SV=1
MM1 pKa = 6.71EE2 pKa = 4.7TLVGGLLTGEE12 pKa = 4.92DD13 pKa = 4.05SLISMSNDD21 pKa = 2.95VSCLYY26 pKa = 10.87VYY28 pKa = 10.04DD29 pKa = 3.75GPMRR33 pKa = 11.84VFSQNALMPTLQSVKK48 pKa = 10.46RR49 pKa = 11.84SDD51 pKa = 3.35QFSKK55 pKa = 11.07GKK57 pKa = 7.77TKK59 pKa = 10.7RR60 pKa = 11.84FIIDD64 pKa = 3.73LFGMKK69 pKa = 10.19RR70 pKa = 11.84MWDD73 pKa = 3.15IGNKK77 pKa = 7.83QLEE80 pKa = 4.44DD81 pKa = 3.73EE82 pKa = 4.77NLDD85 pKa = 3.68EE86 pKa = 4.55TVGVADD92 pKa = 4.99LGLVKK97 pKa = 10.61YY98 pKa = 10.05LINNKK103 pKa = 9.32YY104 pKa = 10.67DD105 pKa = 3.72EE106 pKa = 4.64AEE108 pKa = 4.08KK109 pKa = 10.18TSLRR113 pKa = 11.84KK114 pKa = 10.15SMEE117 pKa = 3.84EE118 pKa = 3.67AFEE121 pKa = 4.32KK122 pKa = 11.0SMNEE126 pKa = 3.68EE127 pKa = 3.51FVVLNKK133 pKa = 10.45GKK135 pKa = 10.03SANDD139 pKa = 3.57IISDD143 pKa = 3.82TNAMCKK149 pKa = 10.23FCVKK153 pKa = 9.84NWIVATGFRR162 pKa = 11.84GRR164 pKa = 11.84TMSDD168 pKa = 3.47LIEE171 pKa = 3.88HH172 pKa = 7.41HH173 pKa = 6.37FRR175 pKa = 11.84CMQGKK180 pKa = 9.06QEE182 pKa = 3.99VKK184 pKa = 10.51GYY186 pKa = 9.05IWKK189 pKa = 9.79HH190 pKa = 5.44KK191 pKa = 9.72YY192 pKa = 9.12NEE194 pKa = 3.93RR195 pKa = 11.84LKK197 pKa = 10.7RR198 pKa = 11.84KK199 pKa = 8.68QLSKK203 pKa = 11.48EE204 pKa = 3.75EE205 pKa = 4.07VKK207 pKa = 10.5FDD209 pKa = 3.54RR210 pKa = 11.84EE211 pKa = 4.49EE212 pKa = 3.89YY213 pKa = 8.45TSRR216 pKa = 11.84SFRR219 pKa = 11.84LLSFLKK225 pKa = 10.52NSEE228 pKa = 4.1RR229 pKa = 11.84TKK231 pKa = 10.83LEE233 pKa = 3.52PRR235 pKa = 11.84AVFTAGVPWRR245 pKa = 11.84AFIFVLEE252 pKa = 4.05QTMLVVNKK260 pKa = 9.88LDD262 pKa = 4.1PNSVIWMGSDD272 pKa = 3.33AKK274 pKa = 10.97INTTNSRR281 pKa = 11.84IKK283 pKa = 10.29EE284 pKa = 3.77IGMKK288 pKa = 10.28NQGQTLVTLTGDD300 pKa = 3.04NSKK303 pKa = 10.79YY304 pKa = 10.18NEE306 pKa = 4.27SMCPEE311 pKa = 3.7VMMVFLRR318 pKa = 11.84EE319 pKa = 4.09LGIKK323 pKa = 10.48GPMLEE328 pKa = 4.2VLDD331 pKa = 3.82YY332 pKa = 11.42ALWQFSQKK340 pKa = 8.96SVKK343 pKa = 9.71PVAPIKK349 pKa = 10.64KK350 pKa = 8.05RR351 pKa = 11.84TGKK354 pKa = 8.28STVVIKK360 pKa = 10.74ADD362 pKa = 3.55SVKK365 pKa = 10.36EE366 pKa = 3.81CRR368 pKa = 11.84DD369 pKa = 3.23AFNEE373 pKa = 4.25KK374 pKa = 9.95EE375 pKa = 4.07LEE377 pKa = 4.54LIQGVEE383 pKa = 3.73WMDD386 pKa = 3.69DD387 pKa = 3.0GFVRR391 pKa = 11.84VRR393 pKa = 11.84RR394 pKa = 11.84GMLMGMANNAFTTASTIASSFSFTPEE420 pKa = 3.34AVYY423 pKa = 9.53TLQSSDD429 pKa = 3.85DD430 pKa = 4.17FVTGSCGRR438 pKa = 11.84DD439 pKa = 3.32VQHH442 pKa = 6.72ARR444 pKa = 11.84QRR446 pKa = 11.84LEE448 pKa = 3.68MALKK452 pKa = 9.45VSKK455 pKa = 10.64AAGLNVSQKK464 pKa = 10.48KK465 pKa = 9.77SFYY468 pKa = 10.8VEE470 pKa = 3.68GTTFEE475 pKa = 4.31FNSMFVRR482 pKa = 11.84DD483 pKa = 4.43GKK485 pKa = 11.47VMANGGNFEE494 pKa = 4.38NMTVPGGLGPSTDD507 pKa = 3.34LFVVGKK513 pKa = 7.41QARR516 pKa = 11.84NSMLRR521 pKa = 11.84GNLSFSQAMEE531 pKa = 3.78MCKK534 pKa = 9.92IGITNVEE541 pKa = 3.68KK542 pKa = 10.78VYY544 pKa = 10.92YY545 pKa = 10.35GNRR548 pKa = 11.84KK549 pKa = 7.82YY550 pKa = 10.61QEE552 pKa = 4.1LKK554 pKa = 10.25NEE556 pKa = 3.71IRR558 pKa = 11.84EE559 pKa = 4.29KK560 pKa = 10.79CGEE563 pKa = 4.03EE564 pKa = 4.05TMSIPEE570 pKa = 4.06SMGGDD575 pKa = 3.35RR576 pKa = 11.84KK577 pKa = 9.75PRR579 pKa = 11.84PWEE582 pKa = 3.91LPQSFDD588 pKa = 4.4GIALKK593 pKa = 10.51EE594 pKa = 4.04AVNRR598 pKa = 11.84GHH600 pKa = 6.93WKK602 pKa = 7.77AAKK605 pKa = 9.64YY606 pKa = 10.07IKK608 pKa = 9.9SCCSIEE614 pKa = 3.84FDD616 pKa = 4.94EE617 pKa = 5.93EE618 pKa = 5.1GDD620 pKa = 4.16QSWDD624 pKa = 3.22TSKK627 pKa = 9.73TALVVIRR634 pKa = 11.84KK635 pKa = 9.35NEE637 pKa = 3.46TDD639 pKa = 2.89MRR641 pKa = 11.84RR642 pKa = 11.84RR643 pKa = 11.84TVKK646 pKa = 9.35TRR648 pKa = 11.84NPKK651 pKa = 10.58DD652 pKa = 3.68KK653 pKa = 10.77IFNDD657 pKa = 3.27AMNKK661 pKa = 8.56AKK663 pKa = 10.38RR664 pKa = 11.84MYY666 pKa = 8.79EE667 pKa = 4.05TVVDD671 pKa = 4.64RR672 pKa = 11.84NPLLGLKK679 pKa = 10.29GKK681 pKa = 10.3GGRR684 pKa = 11.84LTVKK688 pKa = 10.13DD689 pKa = 3.81LKK691 pKa = 10.87ARR693 pKa = 11.84KK694 pKa = 9.55LIDD697 pKa = 3.32EE698 pKa = 4.47VEE700 pKa = 4.3VVKK703 pKa = 10.76KK704 pKa = 10.36KK705 pKa = 10.56KK706 pKa = 9.93HH707 pKa = 3.95VV708 pKa = 3.3
MM1 pKa = 6.71EE2 pKa = 4.7TLVGGLLTGEE12 pKa = 4.92DD13 pKa = 4.05SLISMSNDD21 pKa = 2.95VSCLYY26 pKa = 10.87VYY28 pKa = 10.04DD29 pKa = 3.75GPMRR33 pKa = 11.84VFSQNALMPTLQSVKK48 pKa = 10.46RR49 pKa = 11.84SDD51 pKa = 3.35QFSKK55 pKa = 11.07GKK57 pKa = 7.77TKK59 pKa = 10.7RR60 pKa = 11.84FIIDD64 pKa = 3.73LFGMKK69 pKa = 10.19RR70 pKa = 11.84MWDD73 pKa = 3.15IGNKK77 pKa = 7.83QLEE80 pKa = 4.44DD81 pKa = 3.73EE82 pKa = 4.77NLDD85 pKa = 3.68EE86 pKa = 4.55TVGVADD92 pKa = 4.99LGLVKK97 pKa = 10.61YY98 pKa = 10.05LINNKK103 pKa = 9.32YY104 pKa = 10.67DD105 pKa = 3.72EE106 pKa = 4.64AEE108 pKa = 4.08KK109 pKa = 10.18TSLRR113 pKa = 11.84KK114 pKa = 10.15SMEE117 pKa = 3.84EE118 pKa = 3.67AFEE121 pKa = 4.32KK122 pKa = 11.0SMNEE126 pKa = 3.68EE127 pKa = 3.51FVVLNKK133 pKa = 10.45GKK135 pKa = 10.03SANDD139 pKa = 3.57IISDD143 pKa = 3.82TNAMCKK149 pKa = 10.23FCVKK153 pKa = 9.84NWIVATGFRR162 pKa = 11.84GRR164 pKa = 11.84TMSDD168 pKa = 3.47LIEE171 pKa = 3.88HH172 pKa = 7.41HH173 pKa = 6.37FRR175 pKa = 11.84CMQGKK180 pKa = 9.06QEE182 pKa = 3.99VKK184 pKa = 10.51GYY186 pKa = 9.05IWKK189 pKa = 9.79HH190 pKa = 5.44KK191 pKa = 9.72YY192 pKa = 9.12NEE194 pKa = 3.93RR195 pKa = 11.84LKK197 pKa = 10.7RR198 pKa = 11.84KK199 pKa = 8.68QLSKK203 pKa = 11.48EE204 pKa = 3.75EE205 pKa = 4.07VKK207 pKa = 10.5FDD209 pKa = 3.54RR210 pKa = 11.84EE211 pKa = 4.49EE212 pKa = 3.89YY213 pKa = 8.45TSRR216 pKa = 11.84SFRR219 pKa = 11.84LLSFLKK225 pKa = 10.52NSEE228 pKa = 4.1RR229 pKa = 11.84TKK231 pKa = 10.83LEE233 pKa = 3.52PRR235 pKa = 11.84AVFTAGVPWRR245 pKa = 11.84AFIFVLEE252 pKa = 4.05QTMLVVNKK260 pKa = 9.88LDD262 pKa = 4.1PNSVIWMGSDD272 pKa = 3.33AKK274 pKa = 10.97INTTNSRR281 pKa = 11.84IKK283 pKa = 10.29EE284 pKa = 3.77IGMKK288 pKa = 10.28NQGQTLVTLTGDD300 pKa = 3.04NSKK303 pKa = 10.79YY304 pKa = 10.18NEE306 pKa = 4.27SMCPEE311 pKa = 3.7VMMVFLRR318 pKa = 11.84EE319 pKa = 4.09LGIKK323 pKa = 10.48GPMLEE328 pKa = 4.2VLDD331 pKa = 3.82YY332 pKa = 11.42ALWQFSQKK340 pKa = 8.96SVKK343 pKa = 9.71PVAPIKK349 pKa = 10.64KK350 pKa = 8.05RR351 pKa = 11.84TGKK354 pKa = 8.28STVVIKK360 pKa = 10.74ADD362 pKa = 3.55SVKK365 pKa = 10.36EE366 pKa = 3.81CRR368 pKa = 11.84DD369 pKa = 3.23AFNEE373 pKa = 4.25KK374 pKa = 9.95EE375 pKa = 4.07LEE377 pKa = 4.54LIQGVEE383 pKa = 3.73WMDD386 pKa = 3.69DD387 pKa = 3.0GFVRR391 pKa = 11.84VRR393 pKa = 11.84RR394 pKa = 11.84GMLMGMANNAFTTASTIASSFSFTPEE420 pKa = 3.34AVYY423 pKa = 9.53TLQSSDD429 pKa = 3.85DD430 pKa = 4.17FVTGSCGRR438 pKa = 11.84DD439 pKa = 3.32VQHH442 pKa = 6.72ARR444 pKa = 11.84QRR446 pKa = 11.84LEE448 pKa = 3.68MALKK452 pKa = 9.45VSKK455 pKa = 10.64AAGLNVSQKK464 pKa = 10.48KK465 pKa = 9.77SFYY468 pKa = 10.8VEE470 pKa = 3.68GTTFEE475 pKa = 4.31FNSMFVRR482 pKa = 11.84DD483 pKa = 4.43GKK485 pKa = 11.47VMANGGNFEE494 pKa = 4.38NMTVPGGLGPSTDD507 pKa = 3.34LFVVGKK513 pKa = 7.41QARR516 pKa = 11.84NSMLRR521 pKa = 11.84GNLSFSQAMEE531 pKa = 3.78MCKK534 pKa = 9.92IGITNVEE541 pKa = 3.68KK542 pKa = 10.78VYY544 pKa = 10.92YY545 pKa = 10.35GNRR548 pKa = 11.84KK549 pKa = 7.82YY550 pKa = 10.61QEE552 pKa = 4.1LKK554 pKa = 10.25NEE556 pKa = 3.71IRR558 pKa = 11.84EE559 pKa = 4.29KK560 pKa = 10.79CGEE563 pKa = 4.03EE564 pKa = 4.05TMSIPEE570 pKa = 4.06SMGGDD575 pKa = 3.35RR576 pKa = 11.84KK577 pKa = 9.75PRR579 pKa = 11.84PWEE582 pKa = 3.91LPQSFDD588 pKa = 4.4GIALKK593 pKa = 10.51EE594 pKa = 4.04AVNRR598 pKa = 11.84GHH600 pKa = 6.93WKK602 pKa = 7.77AAKK605 pKa = 9.64YY606 pKa = 10.07IKK608 pKa = 9.9SCCSIEE614 pKa = 3.84FDD616 pKa = 4.94EE617 pKa = 5.93EE618 pKa = 5.1GDD620 pKa = 4.16QSWDD624 pKa = 3.22TSKK627 pKa = 9.73TALVVIRR634 pKa = 11.84KK635 pKa = 9.35NEE637 pKa = 3.46TDD639 pKa = 2.89MRR641 pKa = 11.84RR642 pKa = 11.84RR643 pKa = 11.84TVKK646 pKa = 9.35TRR648 pKa = 11.84NPKK651 pKa = 10.58DD652 pKa = 3.68KK653 pKa = 10.77IFNDD657 pKa = 3.27AMNKK661 pKa = 8.56AKK663 pKa = 10.38RR664 pKa = 11.84MYY666 pKa = 8.79EE667 pKa = 4.05TVVDD671 pKa = 4.64RR672 pKa = 11.84NPLLGLKK679 pKa = 10.29GKK681 pKa = 10.3GGRR684 pKa = 11.84LTVKK688 pKa = 10.13DD689 pKa = 3.81LKK691 pKa = 10.87ARR693 pKa = 11.84KK694 pKa = 9.55LIDD697 pKa = 3.32EE698 pKa = 4.47VEE700 pKa = 4.3VVKK703 pKa = 10.76KK704 pKa = 10.36KK705 pKa = 10.56KK706 pKa = 9.93HH707 pKa = 3.95VV708 pKa = 3.3
Molecular weight: 80.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4281 |
146 |
722 |
428.1 |
47.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.7 ± 0.48 | 2.453 ± 0.353 |
5.466 ± 0.427 | 7.452 ± 0.509 |
3.854 ± 0.363 | 8.339 ± 0.574 |
1.145 ± 0.284 | 4.999 ± 0.451 |
7.381 ± 0.531 | 7.779 ± 0.408 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.018 ± 0.377 | 4.205 ± 0.429 |
3.107 ± 0.279 | 3.2 ± 0.212 |
5.91 ± 0.523 | 7.381 ± 0.48 |
6.657 ± 0.434 | 7.265 ± 0.419 |
1.355 ± 0.171 | 2.219 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
