
Draconibacterium orientale
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Draconibacterium
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
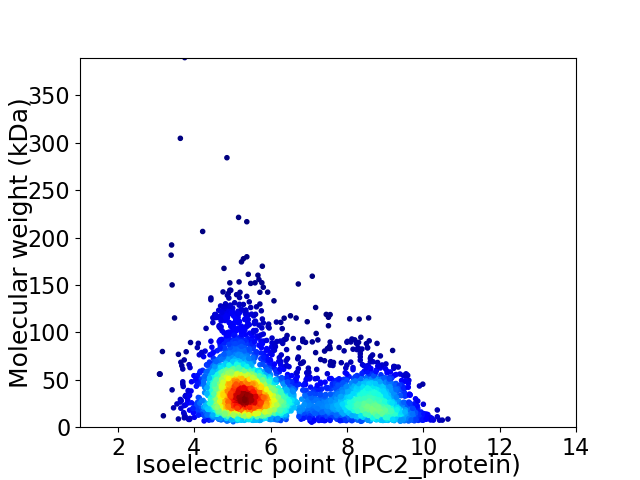
Virtual 2D-PAGE plot for 3940 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|X5DM66|X5DM66_9BACT Hemoglobin/transferrin/lactoferrin receptor protein OS=Draconibacterium orientale OX=1168034 GN=FH5T_06340 PE=3 SV=1
MM1 pKa = 7.61SIYY4 pKa = 10.44GYY6 pKa = 9.38SKK8 pKa = 10.95KK9 pKa = 10.64LVFLFAMVLIAGFVMAQKK27 pKa = 7.67TTTLVQSGNSNIATVDD43 pKa = 3.31QMACLSGAVNSIFATQSGNWNTLTTFQKK71 pKa = 10.81GWDD74 pKa = 3.72NNIDD78 pKa = 3.79LLQSGDD84 pKa = 3.85HH85 pKa = 5.95NTADD89 pKa = 3.88MTQEE93 pKa = 3.75TCDD96 pKa = 3.68YY97 pKa = 10.79DD98 pKa = 4.84DD99 pKa = 3.34SHH101 pKa = 7.9GYY103 pKa = 7.97NTAKK107 pKa = 10.08VVQYY111 pKa = 11.54GDD113 pKa = 3.89GNTANLSQMEE123 pKa = 4.4EE124 pKa = 4.18DD125 pKa = 4.94GGLADD130 pKa = 3.92IDD132 pKa = 4.46YY133 pKa = 11.11VGGWLPLPILPEE145 pKa = 3.96RR146 pKa = 11.84DD147 pKa = 3.1RR148 pKa = 11.84SINEE152 pKa = 3.61ATTTQSGNSNTYY164 pKa = 9.92NLAQGRR170 pKa = 11.84EE171 pKa = 4.02NYY173 pKa = 10.03IPDD176 pKa = 3.36NTSYY180 pKa = 9.83LTQSGNSNAADD191 pKa = 3.26IDD193 pKa = 3.62QLGFNNYY200 pKa = 9.77SEE202 pKa = 4.25IRR204 pKa = 11.84QTGDD208 pKa = 2.37WNTADD213 pKa = 4.7LLQDD217 pKa = 3.67GADD220 pKa = 3.29ISLTAVNKK228 pKa = 10.43DD229 pKa = 3.62EE230 pKa = 4.49SYY232 pKa = 9.05SWQIGVHH239 pKa = 4.61STLNVKK245 pKa = 9.73QVGDD249 pKa = 3.52VDD251 pKa = 3.83EE252 pKa = 4.49QLAEE256 pKa = 4.14SRR258 pKa = 11.84QNGSGNTTNILQVGVTMQNVDD279 pKa = 2.6AWQYY283 pKa = 8.08GTDD286 pKa = 4.08VIDD289 pKa = 3.66IDD291 pKa = 4.02QVDD294 pKa = 4.14VEE296 pKa = 4.46PEE298 pKa = 3.79EE299 pKa = 4.1
MM1 pKa = 7.61SIYY4 pKa = 10.44GYY6 pKa = 9.38SKK8 pKa = 10.95KK9 pKa = 10.64LVFLFAMVLIAGFVMAQKK27 pKa = 7.67TTTLVQSGNSNIATVDD43 pKa = 3.31QMACLSGAVNSIFATQSGNWNTLTTFQKK71 pKa = 10.81GWDD74 pKa = 3.72NNIDD78 pKa = 3.79LLQSGDD84 pKa = 3.85HH85 pKa = 5.95NTADD89 pKa = 3.88MTQEE93 pKa = 3.75TCDD96 pKa = 3.68YY97 pKa = 10.79DD98 pKa = 4.84DD99 pKa = 3.34SHH101 pKa = 7.9GYY103 pKa = 7.97NTAKK107 pKa = 10.08VVQYY111 pKa = 11.54GDD113 pKa = 3.89GNTANLSQMEE123 pKa = 4.4EE124 pKa = 4.18DD125 pKa = 4.94GGLADD130 pKa = 3.92IDD132 pKa = 4.46YY133 pKa = 11.11VGGWLPLPILPEE145 pKa = 3.96RR146 pKa = 11.84DD147 pKa = 3.1RR148 pKa = 11.84SINEE152 pKa = 3.61ATTTQSGNSNTYY164 pKa = 9.92NLAQGRR170 pKa = 11.84EE171 pKa = 4.02NYY173 pKa = 10.03IPDD176 pKa = 3.36NTSYY180 pKa = 9.83LTQSGNSNAADD191 pKa = 3.26IDD193 pKa = 3.62QLGFNNYY200 pKa = 9.77SEE202 pKa = 4.25IRR204 pKa = 11.84QTGDD208 pKa = 2.37WNTADD213 pKa = 4.7LLQDD217 pKa = 3.67GADD220 pKa = 3.29ISLTAVNKK228 pKa = 10.43DD229 pKa = 3.62EE230 pKa = 4.49SYY232 pKa = 9.05SWQIGVHH239 pKa = 4.61STLNVKK245 pKa = 9.73QVGDD249 pKa = 3.52VDD251 pKa = 3.83EE252 pKa = 4.49QLAEE256 pKa = 4.14SRR258 pKa = 11.84QNGSGNTTNILQVGVTMQNVDD279 pKa = 2.6AWQYY283 pKa = 8.08GTDD286 pKa = 4.08VIDD289 pKa = 3.66IDD291 pKa = 4.02QVDD294 pKa = 4.14VEE296 pKa = 4.46PEE298 pKa = 3.79EE299 pKa = 4.1
Molecular weight: 32.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X5DLY7|X5DLY7_9BACT Curlin associated repeat-containing protein OS=Draconibacterium orientale OX=1168034 GN=FH5T_18445 PE=3 SV=1
MM1 pKa = 7.6LRR3 pKa = 11.84TDD5 pKa = 3.53CRR7 pKa = 11.84KK8 pKa = 10.16LKK10 pKa = 10.65NSNQRR15 pKa = 11.84KK16 pKa = 8.04QLILNLPQVPEE27 pKa = 3.98VSFKK31 pKa = 10.88RR32 pKa = 11.84VFLRR36 pKa = 11.84PQLTEE41 pKa = 3.66NGFKK45 pKa = 10.33RR46 pKa = 11.84VSLLHH51 pKa = 5.64QVTEE55 pKa = 4.34EE56 pKa = 4.0GFKK59 pKa = 10.68RR60 pKa = 11.84IFARR64 pKa = 11.84RR65 pKa = 11.84QINRR69 pKa = 11.84VDD71 pKa = 3.36TKK73 pKa = 10.94RR74 pKa = 11.84IFSCAQHH81 pKa = 6.19IAGSTKK87 pKa = 10.3HH88 pKa = 6.51ILTGHH93 pKa = 5.61CHH95 pKa = 5.64MGIKK99 pKa = 10.19NISFYY104 pKa = 11.34SVVIQKK110 pKa = 8.54TGNSHH115 pKa = 6.82VEE117 pKa = 3.32ICYY120 pKa = 10.21LRR122 pKa = 11.84SGPFFTLAYY131 pKa = 10.27FMAKK135 pKa = 9.74NVV137 pKa = 3.29
MM1 pKa = 7.6LRR3 pKa = 11.84TDD5 pKa = 3.53CRR7 pKa = 11.84KK8 pKa = 10.16LKK10 pKa = 10.65NSNQRR15 pKa = 11.84KK16 pKa = 8.04QLILNLPQVPEE27 pKa = 3.98VSFKK31 pKa = 10.88RR32 pKa = 11.84VFLRR36 pKa = 11.84PQLTEE41 pKa = 3.66NGFKK45 pKa = 10.33RR46 pKa = 11.84VSLLHH51 pKa = 5.64QVTEE55 pKa = 4.34EE56 pKa = 4.0GFKK59 pKa = 10.68RR60 pKa = 11.84IFARR64 pKa = 11.84RR65 pKa = 11.84QINRR69 pKa = 11.84VDD71 pKa = 3.36TKK73 pKa = 10.94RR74 pKa = 11.84IFSCAQHH81 pKa = 6.19IAGSTKK87 pKa = 10.3HH88 pKa = 6.51ILTGHH93 pKa = 5.61CHH95 pKa = 5.64MGIKK99 pKa = 10.19NISFYY104 pKa = 11.34SVVIQKK110 pKa = 8.54TGNSHH115 pKa = 6.82VEE117 pKa = 3.32ICYY120 pKa = 10.21LRR122 pKa = 11.84SGPFFTLAYY131 pKa = 10.27FMAKK135 pKa = 9.74NVV137 pKa = 3.29
Molecular weight: 15.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1407688 |
48 |
3688 |
357.3 |
40.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.891 ± 0.034 | 0.844 ± 0.016 |
5.585 ± 0.033 | 7.037 ± 0.037 |
5.064 ± 0.028 | 6.758 ± 0.04 |
1.806 ± 0.015 | 7.275 ± 0.039 |
7.035 ± 0.047 | 9.135 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.411 ± 0.018 | 5.702 ± 0.031 |
3.662 ± 0.023 | 3.504 ± 0.025 |
3.872 ± 0.024 | 6.091 ± 0.028 |
5.438 ± 0.04 | 6.544 ± 0.028 |
1.271 ± 0.016 | 4.074 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
