
Faeces associated gemycircularvirus 16
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus sheas1; Sheep associated gemycircularvirus 1
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
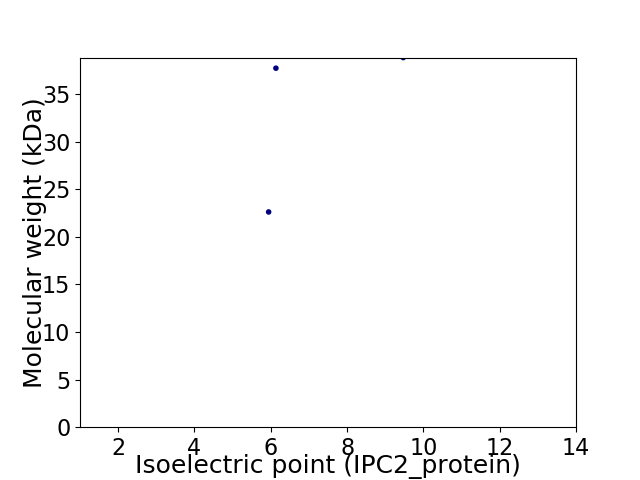
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A160HWN0|A0A160HWN0_9VIRU Replication-associated protein OS=Faeces associated gemycircularvirus 16 OX=1843736 PE=3 SV=1
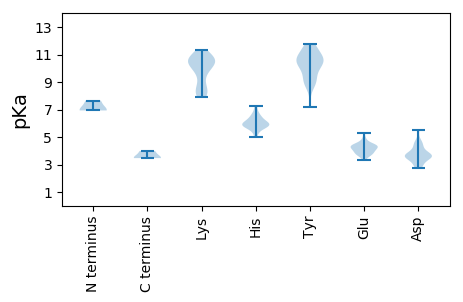
MM1 pKa = 6.93TQMLFCNSKK10 pKa = 10.64YY11 pKa = 11.1VLLTYY16 pKa = 9.13AQCGDD21 pKa = 3.67LDD23 pKa = 3.59EE24 pKa = 4.86WAVSDD29 pKa = 4.21HH30 pKa = 6.46LSSLGAEE37 pKa = 4.23CIVARR42 pKa = 11.84EE43 pKa = 4.04LHH45 pKa = 6.0PTTGGVHH52 pKa = 5.86LHH54 pKa = 5.79VFVDD58 pKa = 4.54FGRR61 pKa = 11.84KK62 pKa = 7.9FRR64 pKa = 11.84SRR66 pKa = 11.84RR67 pKa = 11.84VDD69 pKa = 2.87IFDD72 pKa = 3.52VEE74 pKa = 4.44GRR76 pKa = 11.84HH77 pKa = 5.97PNVVPSKK84 pKa = 8.26GTPEE88 pKa = 3.8KK89 pKa = 10.85GYY91 pKa = 10.88DD92 pKa = 3.43YY93 pKa = 10.61AIKK96 pKa = 10.61DD97 pKa = 3.82GEE99 pKa = 4.57VVAGGLEE106 pKa = 4.08RR107 pKa = 11.84PRR109 pKa = 11.84GGRR112 pKa = 11.84PGTVSGLEE120 pKa = 4.4AIAHH124 pKa = 5.74LCEE127 pKa = 4.35TQDD130 pKa = 3.61EE131 pKa = 4.36FLDD134 pKa = 3.72IYY136 pKa = 11.48GEE138 pKa = 3.96VDD140 pKa = 2.94TRR142 pKa = 11.84GLIKK146 pKa = 10.77NFANVRR152 pKa = 11.84SYY154 pKa = 11.77AKK156 pKa = 9.08WRR158 pKa = 11.84YY159 pKa = 10.07AGTLPKK165 pKa = 10.45YY166 pKa = 9.07EE167 pKa = 4.12SPYY170 pKa = 10.52SIGEE174 pKa = 3.78FRR176 pKa = 11.84RR177 pKa = 11.84GSDD180 pKa = 5.52GRR182 pKa = 11.84DD183 pKa = 2.71QWLAQSGIRR192 pKa = 11.84SGDD195 pKa = 3.58LVVSKK200 pKa = 10.33CFQQ203 pKa = 3.47
MM1 pKa = 6.93TQMLFCNSKK10 pKa = 10.64YY11 pKa = 11.1VLLTYY16 pKa = 9.13AQCGDD21 pKa = 3.67LDD23 pKa = 3.59EE24 pKa = 4.86WAVSDD29 pKa = 4.21HH30 pKa = 6.46LSSLGAEE37 pKa = 4.23CIVARR42 pKa = 11.84EE43 pKa = 4.04LHH45 pKa = 6.0PTTGGVHH52 pKa = 5.86LHH54 pKa = 5.79VFVDD58 pKa = 4.54FGRR61 pKa = 11.84KK62 pKa = 7.9FRR64 pKa = 11.84SRR66 pKa = 11.84RR67 pKa = 11.84VDD69 pKa = 2.87IFDD72 pKa = 3.52VEE74 pKa = 4.44GRR76 pKa = 11.84HH77 pKa = 5.97PNVVPSKK84 pKa = 8.26GTPEE88 pKa = 3.8KK89 pKa = 10.85GYY91 pKa = 10.88DD92 pKa = 3.43YY93 pKa = 10.61AIKK96 pKa = 10.61DD97 pKa = 3.82GEE99 pKa = 4.57VVAGGLEE106 pKa = 4.08RR107 pKa = 11.84PRR109 pKa = 11.84GGRR112 pKa = 11.84PGTVSGLEE120 pKa = 4.4AIAHH124 pKa = 5.74LCEE127 pKa = 4.35TQDD130 pKa = 3.61EE131 pKa = 4.36FLDD134 pKa = 3.72IYY136 pKa = 11.48GEE138 pKa = 3.96VDD140 pKa = 2.94TRR142 pKa = 11.84GLIKK146 pKa = 10.77NFANVRR152 pKa = 11.84SYY154 pKa = 11.77AKK156 pKa = 9.08WRR158 pKa = 11.84YY159 pKa = 10.07AGTLPKK165 pKa = 10.45YY166 pKa = 9.07EE167 pKa = 4.12SPYY170 pKa = 10.52SIGEE174 pKa = 3.78FRR176 pKa = 11.84RR177 pKa = 11.84GSDD180 pKa = 5.52GRR182 pKa = 11.84DD183 pKa = 2.71QWLAQSGIRR192 pKa = 11.84SGDD195 pKa = 3.58LVVSKK200 pKa = 10.33CFQQ203 pKa = 3.47
Molecular weight: 22.61 kDa
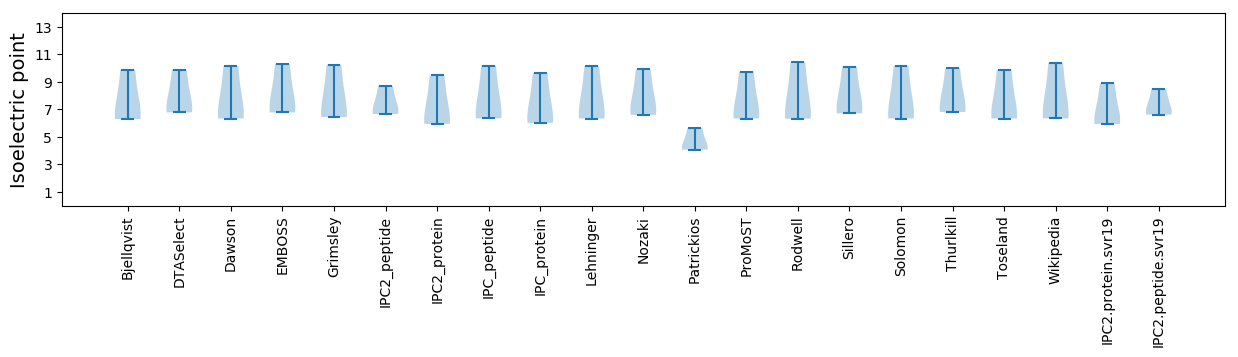
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MG69|A0A168MG69_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 16 OX=1843736 PE=4 SV=1
MM1 pKa = 7.65PRR3 pKa = 11.84FKK5 pKa = 10.87NRR7 pKa = 11.84RR8 pKa = 11.84TRR10 pKa = 11.84FARR13 pKa = 11.84KK14 pKa = 8.7AKK16 pKa = 9.69VWFKK20 pKa = 11.1EE21 pKa = 3.54KK22 pKa = 10.55LPRR25 pKa = 11.84STRR28 pKa = 11.84RR29 pKa = 11.84VTKK32 pKa = 8.3RR33 pKa = 11.84TYY35 pKa = 9.6RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84PMTKK43 pKa = 9.84EE44 pKa = 3.59VDD46 pKa = 3.58PQPQRR51 pKa = 11.84RR52 pKa = 11.84SRR54 pKa = 11.84KK55 pKa = 8.15KK56 pKa = 8.5GCHH59 pKa = 5.87ASVTNPTLDD68 pKa = 3.68PLIASSASPAILGSGRR84 pKa = 11.84PAPSGAPAYY93 pKa = 7.21TYY95 pKa = 9.09CIPWIATRR103 pKa = 11.84RR104 pKa = 11.84KK105 pKa = 8.0PANVGNSAMAQLGTGTPYY123 pKa = 10.31MKK125 pKa = 10.55GLSEE129 pKa = 5.29NITISTTGAAPWQWRR144 pKa = 11.84RR145 pKa = 11.84VCFTFKK151 pKa = 10.44SKK153 pKa = 11.16YY154 pKa = 10.26LIEE157 pKa = 4.3QFTISQGDD165 pKa = 3.68DD166 pKa = 3.9LIDD169 pKa = 4.34FNHH172 pKa = 7.29DD173 pKa = 3.44PFFSNGYY180 pKa = 8.6NDD182 pKa = 3.07SSGVFRR188 pKa = 11.84PMYY191 pKa = 10.53DD192 pKa = 2.97LASYY196 pKa = 9.53YY197 pKa = 10.78GKK199 pKa = 10.11HH200 pKa = 5.66NNPLKK205 pKa = 10.54FRR207 pKa = 11.84ARR209 pKa = 11.84SPQHH213 pKa = 5.33TLGFSTLCSVVAAQQLPGQPTQADD237 pKa = 4.57YY238 pKa = 11.49INVMTAKK245 pKa = 9.69TDD247 pKa = 3.54NTEE250 pKa = 3.74VTIKK254 pKa = 10.45YY255 pKa = 9.9DD256 pKa = 3.34KK257 pKa = 10.56TVTIASGNEE266 pKa = 3.31AGVQRR271 pKa = 11.84NYY273 pKa = 10.6RR274 pKa = 11.84RR275 pKa = 11.84YY276 pKa = 9.74HH277 pKa = 6.11PMNKK281 pKa = 8.41TLVYY285 pKa = 10.71DD286 pKa = 3.77SLEE289 pKa = 3.9QGSNSNYY296 pKa = 10.58DD297 pKa = 3.49SMSTEE302 pKa = 4.37AKK304 pKa = 10.09PGMGDD309 pKa = 3.59YY310 pKa = 10.92YY311 pKa = 11.56VVDD314 pKa = 4.59FFQARR319 pKa = 11.84YY320 pKa = 8.86LADD323 pKa = 4.58ADD325 pKa = 4.18DD326 pKa = 4.88GNLIFDD332 pKa = 4.66PRR334 pKa = 11.84ATLYY338 pKa = 8.62WHH340 pKa = 6.56EE341 pKa = 4.23RR342 pKa = 3.47
MM1 pKa = 7.65PRR3 pKa = 11.84FKK5 pKa = 10.87NRR7 pKa = 11.84RR8 pKa = 11.84TRR10 pKa = 11.84FARR13 pKa = 11.84KK14 pKa = 8.7AKK16 pKa = 9.69VWFKK20 pKa = 11.1EE21 pKa = 3.54KK22 pKa = 10.55LPRR25 pKa = 11.84STRR28 pKa = 11.84RR29 pKa = 11.84VTKK32 pKa = 8.3RR33 pKa = 11.84TYY35 pKa = 9.6RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84PMTKK43 pKa = 9.84EE44 pKa = 3.59VDD46 pKa = 3.58PQPQRR51 pKa = 11.84RR52 pKa = 11.84SRR54 pKa = 11.84KK55 pKa = 8.15KK56 pKa = 8.5GCHH59 pKa = 5.87ASVTNPTLDD68 pKa = 3.68PLIASSASPAILGSGRR84 pKa = 11.84PAPSGAPAYY93 pKa = 7.21TYY95 pKa = 9.09CIPWIATRR103 pKa = 11.84RR104 pKa = 11.84KK105 pKa = 8.0PANVGNSAMAQLGTGTPYY123 pKa = 10.31MKK125 pKa = 10.55GLSEE129 pKa = 5.29NITISTTGAAPWQWRR144 pKa = 11.84RR145 pKa = 11.84VCFTFKK151 pKa = 10.44SKK153 pKa = 11.16YY154 pKa = 10.26LIEE157 pKa = 4.3QFTISQGDD165 pKa = 3.68DD166 pKa = 3.9LIDD169 pKa = 4.34FNHH172 pKa = 7.29DD173 pKa = 3.44PFFSNGYY180 pKa = 8.6NDD182 pKa = 3.07SSGVFRR188 pKa = 11.84PMYY191 pKa = 10.53DD192 pKa = 2.97LASYY196 pKa = 9.53YY197 pKa = 10.78GKK199 pKa = 10.11HH200 pKa = 5.66NNPLKK205 pKa = 10.54FRR207 pKa = 11.84ARR209 pKa = 11.84SPQHH213 pKa = 5.33TLGFSTLCSVVAAQQLPGQPTQADD237 pKa = 4.57YY238 pKa = 11.49INVMTAKK245 pKa = 9.69TDD247 pKa = 3.54NTEE250 pKa = 3.74VTIKK254 pKa = 10.45YY255 pKa = 9.9DD256 pKa = 3.34KK257 pKa = 10.56TVTIASGNEE266 pKa = 3.31AGVQRR271 pKa = 11.84NYY273 pKa = 10.6RR274 pKa = 11.84RR275 pKa = 11.84YY276 pKa = 9.74HH277 pKa = 6.11PMNKK281 pKa = 8.41TLVYY285 pKa = 10.71DD286 pKa = 3.77SLEE289 pKa = 3.9QGSNSNYY296 pKa = 10.58DD297 pKa = 3.49SMSTEE302 pKa = 4.37AKK304 pKa = 10.09PGMGDD309 pKa = 3.59YY310 pKa = 10.92YY311 pKa = 11.56VVDD314 pKa = 4.59FFQARR319 pKa = 11.84YY320 pKa = 8.86LADD323 pKa = 4.58ADD325 pKa = 4.18DD326 pKa = 4.88GNLIFDD332 pKa = 4.66PRR334 pKa = 11.84ATLYY338 pKa = 8.62WHH340 pKa = 6.56EE341 pKa = 4.23RR342 pKa = 3.47
Molecular weight: 38.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
881 |
203 |
342 |
293.7 |
33.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.151 ± 0.545 | 1.93 ± 0.369 |
6.47 ± 0.447 | 4.654 ± 0.995 |
4.881 ± 0.368 | 9.194 ± 1.483 |
2.497 ± 0.36 | 3.973 ± 0.098 |
5.108 ± 0.367 | 6.583 ± 0.662 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.93 ± 0.386 | 3.519 ± 0.861 |
5.335 ± 0.825 | 3.065 ± 0.538 |
8.059 ± 0.36 | 6.924 ± 0.361 |
5.675 ± 1.226 | 6.47 ± 0.913 |
1.93 ± 0.356 | 4.654 ± 0.456 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
