
Sweet potato leaf curl virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 7.8
Get precalculated fractions of proteins
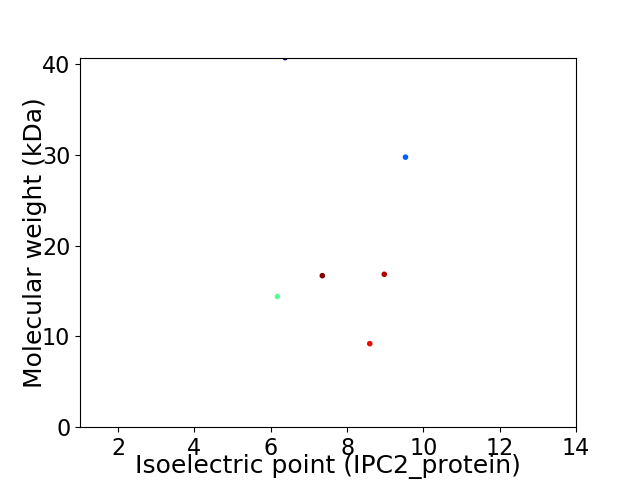
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
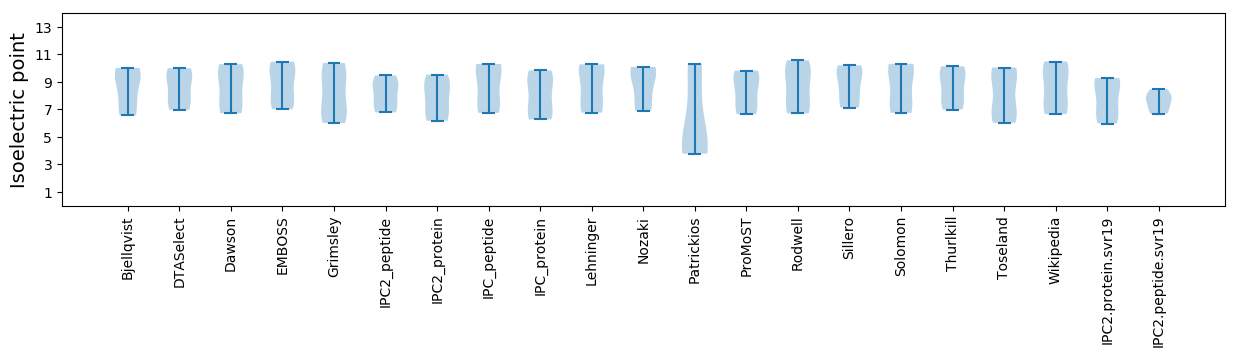
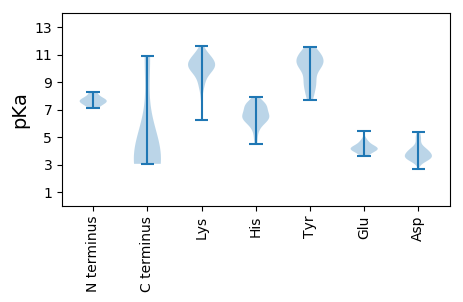
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9PED5|J9PED5_9GEMI Replication-associated protein OS=Sweet potato leaf curl virus OX=100755 PE=3 SV=1
MM1 pKa = 7.68LFQSWSPSMADD12 pKa = 2.69MWDD15 pKa = 3.63PLQNPLPDD23 pKa = 3.26TLYY26 pKa = 11.06GFRR29 pKa = 11.84CMLSVKK35 pKa = 9.89YY36 pKa = 10.19LQSILKK42 pKa = 9.68KK43 pKa = 10.31YY44 pKa = 10.47EE45 pKa = 4.14PGTLGFEE52 pKa = 4.51LCSEE56 pKa = 5.38LIRR59 pKa = 11.84IFRR62 pKa = 11.84VRR64 pKa = 11.84QYY66 pKa = 11.53DD67 pKa = 3.6RR68 pKa = 11.84ANARR72 pKa = 11.84YY73 pKa = 9.98AEE75 pKa = 4.24ISSVWGEE82 pKa = 4.06TGKK85 pKa = 8.61TEE87 pKa = 4.27AEE89 pKa = 3.93LRR91 pKa = 11.84DD92 pKa = 4.14SYY94 pKa = 11.36RR95 pKa = 11.84ALHH98 pKa = 6.22WEE100 pKa = 4.38CCPNCCPKK108 pKa = 10.53LCPGFKK114 pKa = 9.88RR115 pKa = 11.84RR116 pKa = 11.84PDD118 pKa = 3.47EE119 pKa = 4.28EE120 pKa = 4.75KK121 pKa = 10.81EE122 pKa = 4.18GG123 pKa = 3.68
MM1 pKa = 7.68LFQSWSPSMADD12 pKa = 2.69MWDD15 pKa = 3.63PLQNPLPDD23 pKa = 3.26TLYY26 pKa = 11.06GFRR29 pKa = 11.84CMLSVKK35 pKa = 9.89YY36 pKa = 10.19LQSILKK42 pKa = 9.68KK43 pKa = 10.31YY44 pKa = 10.47EE45 pKa = 4.14PGTLGFEE52 pKa = 4.51LCSEE56 pKa = 5.38LIRR59 pKa = 11.84IFRR62 pKa = 11.84VRR64 pKa = 11.84QYY66 pKa = 11.53DD67 pKa = 3.6RR68 pKa = 11.84ANARR72 pKa = 11.84YY73 pKa = 9.98AEE75 pKa = 4.24ISSVWGEE82 pKa = 4.06TGKK85 pKa = 8.61TEE87 pKa = 4.27AEE89 pKa = 3.93LRR91 pKa = 11.84DD92 pKa = 4.14SYY94 pKa = 11.36RR95 pKa = 11.84ALHH98 pKa = 6.22WEE100 pKa = 4.38CCPNCCPKK108 pKa = 10.53LCPGFKK114 pKa = 9.88RR115 pKa = 11.84RR116 pKa = 11.84PDD118 pKa = 3.47EE119 pKa = 4.28EE120 pKa = 4.75KK121 pKa = 10.81EE122 pKa = 4.18GG123 pKa = 3.68
Molecular weight: 14.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9PEW5|J9PEW5_9GEMI Replication enhancer OS=Sweet potato leaf curl virus OX=100755 PE=3 SV=1
MM1 pKa = 7.12TGRR4 pKa = 11.84MRR6 pKa = 11.84VTQRR10 pKa = 11.84FHH12 pKa = 7.35PYY14 pKa = 8.86GGRR17 pKa = 11.84PVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84LNFEE26 pKa = 3.93TAIVPYY32 pKa = 9.22TGNAVPIAARR42 pKa = 11.84SYY44 pKa = 8.64VPVSRR49 pKa = 11.84GVRR52 pKa = 11.84MKK54 pKa = 10.51RR55 pKa = 11.84KK56 pKa = 9.94RR57 pKa = 11.84GDD59 pKa = 4.09RR60 pKa = 11.84IPKK63 pKa = 9.95GCVGPCKK70 pKa = 10.08VQDD73 pKa = 3.86YY74 pKa = 8.72EE75 pKa = 4.55FKK77 pKa = 10.1MDD79 pKa = 3.29VPHH82 pKa = 7.25RR83 pKa = 11.84RR84 pKa = 11.84TFVCYY89 pKa = 10.46SDD91 pKa = 3.63FTRR94 pKa = 11.84GTGLTHH100 pKa = 7.28RR101 pKa = 11.84LGKK104 pKa = 9.73RR105 pKa = 11.84VCVKK109 pKa = 10.91SMGIDD114 pKa = 3.27GKK116 pKa = 11.25VWMDD120 pKa = 3.75DD121 pKa = 3.28NVAKK125 pKa = 10.24RR126 pKa = 11.84DD127 pKa = 3.48HH128 pKa = 6.56TNIITYY134 pKa = 9.02WLIRR138 pKa = 11.84DD139 pKa = 3.65RR140 pKa = 11.84RR141 pKa = 11.84PNKK144 pKa = 10.37DD145 pKa = 3.4PLNFDD150 pKa = 3.79QVFTMYY156 pKa = 11.02DD157 pKa = 3.62NEE159 pKa = 4.13PTTAKK164 pKa = 10.17IRR166 pKa = 11.84MDD168 pKa = 3.48LRR170 pKa = 11.84DD171 pKa = 3.42RR172 pKa = 11.84MQVLKK177 pKa = 10.39KK178 pKa = 10.29FSVTVSGGPYY188 pKa = 8.51NHH190 pKa = 7.45KK191 pKa = 10.06EE192 pKa = 3.61QALVRR197 pKa = 11.84KK198 pKa = 8.79FFKK201 pKa = 10.63GLYY204 pKa = 8.96NHH206 pKa = 5.46VTYY209 pKa = 10.88NHH211 pKa = 6.79KK212 pKa = 10.88EE213 pKa = 3.66EE214 pKa = 4.76ANYY217 pKa = 10.32EE218 pKa = 4.16NQLEE222 pKa = 4.28NALMLYY228 pKa = 9.88SASSHH233 pKa = 6.23ASNPVYY239 pKa = 9.35QTLRR243 pKa = 11.84CRR245 pKa = 11.84VYY247 pKa = 10.7FYY249 pKa = 10.92DD250 pKa = 3.57SHH252 pKa = 7.91NNN254 pKa = 3.3
MM1 pKa = 7.12TGRR4 pKa = 11.84MRR6 pKa = 11.84VTQRR10 pKa = 11.84FHH12 pKa = 7.35PYY14 pKa = 8.86GGRR17 pKa = 11.84PVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84LNFEE26 pKa = 3.93TAIVPYY32 pKa = 9.22TGNAVPIAARR42 pKa = 11.84SYY44 pKa = 8.64VPVSRR49 pKa = 11.84GVRR52 pKa = 11.84MKK54 pKa = 10.51RR55 pKa = 11.84KK56 pKa = 9.94RR57 pKa = 11.84GDD59 pKa = 4.09RR60 pKa = 11.84IPKK63 pKa = 9.95GCVGPCKK70 pKa = 10.08VQDD73 pKa = 3.86YY74 pKa = 8.72EE75 pKa = 4.55FKK77 pKa = 10.1MDD79 pKa = 3.29VPHH82 pKa = 7.25RR83 pKa = 11.84RR84 pKa = 11.84TFVCYY89 pKa = 10.46SDD91 pKa = 3.63FTRR94 pKa = 11.84GTGLTHH100 pKa = 7.28RR101 pKa = 11.84LGKK104 pKa = 9.73RR105 pKa = 11.84VCVKK109 pKa = 10.91SMGIDD114 pKa = 3.27GKK116 pKa = 11.25VWMDD120 pKa = 3.75DD121 pKa = 3.28NVAKK125 pKa = 10.24RR126 pKa = 11.84DD127 pKa = 3.48HH128 pKa = 6.56TNIITYY134 pKa = 9.02WLIRR138 pKa = 11.84DD139 pKa = 3.65RR140 pKa = 11.84RR141 pKa = 11.84PNKK144 pKa = 10.37DD145 pKa = 3.4PLNFDD150 pKa = 3.79QVFTMYY156 pKa = 11.02DD157 pKa = 3.62NEE159 pKa = 4.13PTTAKK164 pKa = 10.17IRR166 pKa = 11.84MDD168 pKa = 3.48LRR170 pKa = 11.84DD171 pKa = 3.42RR172 pKa = 11.84MQVLKK177 pKa = 10.39KK178 pKa = 10.29FSVTVSGGPYY188 pKa = 8.51NHH190 pKa = 7.45KK191 pKa = 10.06EE192 pKa = 3.61QALVRR197 pKa = 11.84KK198 pKa = 8.79FFKK201 pKa = 10.63GLYY204 pKa = 8.96NHH206 pKa = 5.46VTYY209 pKa = 10.88NHH211 pKa = 6.79KK212 pKa = 10.88EE213 pKa = 3.66EE214 pKa = 4.76ANYY217 pKa = 10.32EE218 pKa = 4.16NQLEE222 pKa = 4.28NALMLYY228 pKa = 9.88SASSHH233 pKa = 6.23ASNPVYY239 pKa = 9.35QTLRR243 pKa = 11.84CRR245 pKa = 11.84VYY247 pKa = 10.7FYY249 pKa = 10.92DD250 pKa = 3.57SHH252 pKa = 7.91NNN254 pKa = 3.3
Molecular weight: 29.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1118 |
85 |
364 |
186.3 |
21.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.098 ± 0.873 | 2.952 ± 0.781 |
5.098 ± 0.381 | 5.098 ± 0.721 |
4.114 ± 0.563 | 6.172 ± 0.259 |
2.952 ± 0.426 | 5.009 ± 0.638 |
5.725 ± 0.785 | 7.424 ± 1.127 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.604 | 4.919 ± 0.522 |
6.44 ± 0.951 | 4.204 ± 0.649 |
7.424 ± 1.257 | 8.05 ± 1.41 |
5.903 ± 1.127 | 5.546 ± 1.02 |
1.878 ± 0.316 | 3.667 ± 0.683 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
