
Microviridae sp. ctnrr37
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
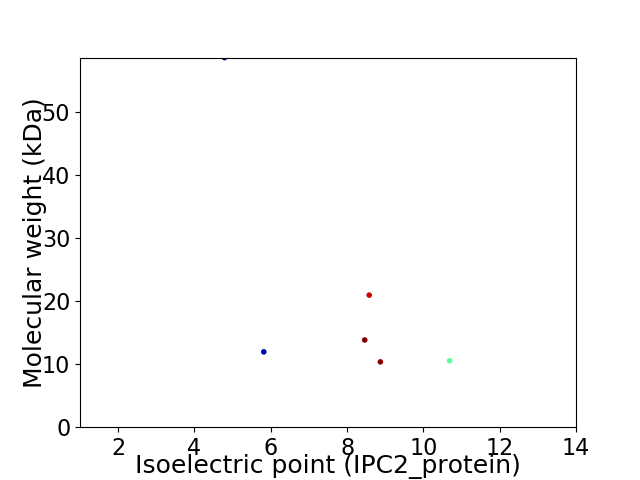
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
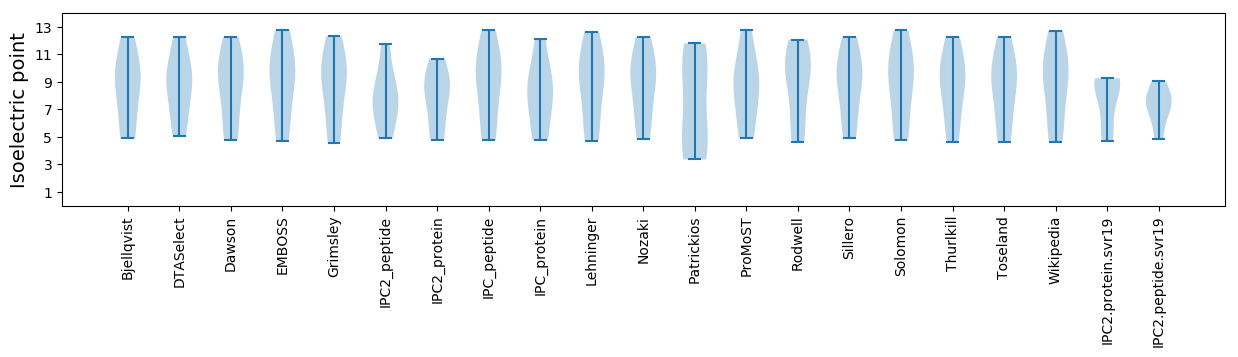
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W727|A0A5Q2W727_9VIRU Scaffold protein OS=Microviridae sp. ctnrr37 OX=2656704 PE=4 SV=1
MM1 pKa = 7.44LRR3 pKa = 11.84GSVPSVSSRR12 pKa = 11.84SQHH15 pKa = 4.63TFSRR19 pKa = 11.84APQAGIPRR27 pKa = 11.84SSFNRR32 pKa = 11.84TFSVKK37 pKa = 7.06TTFNEE42 pKa = 5.29GYY44 pKa = 9.78LVPILADD51 pKa = 3.32EE52 pKa = 4.46MLPGDD57 pKa = 3.88TFSLKK62 pKa = 8.49MHH64 pKa = 6.67AFARR68 pKa = 11.84ISTLLFPIMDD78 pKa = 4.48NLYY81 pKa = 8.88MEE83 pKa = 4.9SFFFFVPNRR92 pKa = 11.84LTWDD96 pKa = 2.84NWEE99 pKa = 4.33NFCQVSTGPTDD110 pKa = 3.33TTEE113 pKa = 3.93YY114 pKa = 10.34TIPQQTGPDD123 pKa = 3.65TMAGGYY129 pKa = 10.49AEE131 pKa = 5.61GSLADD136 pKa = 3.82YY137 pKa = 10.6FGIPTKK143 pKa = 10.55IGGLTPNALHH153 pKa = 6.2FRR155 pKa = 11.84AYY157 pKa = 10.71NLIWNEE163 pKa = 3.75WFRR166 pKa = 11.84SEE168 pKa = 5.07DD169 pKa = 3.33LQAEE173 pKa = 4.62VTVDD177 pKa = 3.32MGDD180 pKa = 3.98GPDD183 pKa = 3.83DD184 pKa = 3.65ATDD187 pKa = 3.59YY188 pKa = 11.64VLLQRR193 pKa = 11.84GKK195 pKa = 10.04RR196 pKa = 11.84YY197 pKa = 10.49DD198 pKa = 3.66YY199 pKa = 8.68FTSCLPWPQKK209 pKa = 11.04GPDD212 pKa = 3.36VLLPLGSSAPVQLISPATDD231 pKa = 2.85TMIGVTGSGNTPVLSQTAIGTNGTGGLQSAPGGNGINIDD270 pKa = 3.93PNGQLIADD278 pKa = 4.39LTDD281 pKa = 3.12ATAATVNQVRR291 pKa = 11.84LAFQTQKK298 pKa = 10.58FYY300 pKa = 11.35EE301 pKa = 4.17KK302 pKa = 9.84QARR305 pKa = 11.84GGTRR309 pKa = 11.84YY310 pKa = 8.49TEE312 pKa = 4.33VIKK315 pKa = 10.96SHH317 pKa = 6.92FDD319 pKa = 3.35VVSPDD324 pKa = 3.11MRR326 pKa = 11.84LQRR329 pKa = 11.84PEE331 pKa = 4.1YY332 pKa = 9.65IGGGSTPVMINPVAQTAPTSGSNALAQLGAYY363 pKa = 7.44GVCAPEE369 pKa = 3.85AHH371 pKa = 6.5GFTYY375 pKa = 10.53SATEE379 pKa = 3.67HH380 pKa = 5.9GVIVGLINVRR390 pKa = 11.84ADD392 pKa = 3.43LNYY395 pKa = 10.4QSGLDD400 pKa = 3.81RR401 pKa = 11.84MWTRR405 pKa = 11.84QVFTDD410 pKa = 3.84YY411 pKa = 9.81YY412 pKa = 9.27WPVFSHH418 pKa = 6.89LGEE421 pKa = 3.99QAVLNQEE428 pKa = 4.66IYY430 pKa = 10.92AQGTSADD437 pKa = 3.65TDD439 pKa = 3.54VFGYY443 pKa = 7.41QEE445 pKa = 3.49RR446 pKa = 11.84HH447 pKa = 4.66AEE449 pKa = 3.95YY450 pKa = 9.88RR451 pKa = 11.84YY452 pKa = 10.22KK453 pKa = 10.72PSLITGLFRR462 pKa = 11.84SNATGTLDD470 pKa = 3.44SWHH473 pKa = 6.89LAQDD477 pKa = 4.29FGSLPEE483 pKa = 4.89LNDD486 pKa = 3.27EE487 pKa = 5.46FIVEE491 pKa = 4.52DD492 pKa = 4.1APMSRR497 pKa = 11.84IKK499 pKa = 10.7AVASSYY505 pKa = 11.59DD506 pKa = 3.43FLFDD510 pKa = 4.12AVFAYY515 pKa = 10.2HH516 pKa = 7.24CARR519 pKa = 11.84PMPVYY524 pKa = 10.31AVPGLIDD531 pKa = 3.33HH532 pKa = 7.15FF533 pKa = 4.9
MM1 pKa = 7.44LRR3 pKa = 11.84GSVPSVSSRR12 pKa = 11.84SQHH15 pKa = 4.63TFSRR19 pKa = 11.84APQAGIPRR27 pKa = 11.84SSFNRR32 pKa = 11.84TFSVKK37 pKa = 7.06TTFNEE42 pKa = 5.29GYY44 pKa = 9.78LVPILADD51 pKa = 3.32EE52 pKa = 4.46MLPGDD57 pKa = 3.88TFSLKK62 pKa = 8.49MHH64 pKa = 6.67AFARR68 pKa = 11.84ISTLLFPIMDD78 pKa = 4.48NLYY81 pKa = 8.88MEE83 pKa = 4.9SFFFFVPNRR92 pKa = 11.84LTWDD96 pKa = 2.84NWEE99 pKa = 4.33NFCQVSTGPTDD110 pKa = 3.33TTEE113 pKa = 3.93YY114 pKa = 10.34TIPQQTGPDD123 pKa = 3.65TMAGGYY129 pKa = 10.49AEE131 pKa = 5.61GSLADD136 pKa = 3.82YY137 pKa = 10.6FGIPTKK143 pKa = 10.55IGGLTPNALHH153 pKa = 6.2FRR155 pKa = 11.84AYY157 pKa = 10.71NLIWNEE163 pKa = 3.75WFRR166 pKa = 11.84SEE168 pKa = 5.07DD169 pKa = 3.33LQAEE173 pKa = 4.62VTVDD177 pKa = 3.32MGDD180 pKa = 3.98GPDD183 pKa = 3.83DD184 pKa = 3.65ATDD187 pKa = 3.59YY188 pKa = 11.64VLLQRR193 pKa = 11.84GKK195 pKa = 10.04RR196 pKa = 11.84YY197 pKa = 10.49DD198 pKa = 3.66YY199 pKa = 8.68FTSCLPWPQKK209 pKa = 11.04GPDD212 pKa = 3.36VLLPLGSSAPVQLISPATDD231 pKa = 2.85TMIGVTGSGNTPVLSQTAIGTNGTGGLQSAPGGNGINIDD270 pKa = 3.93PNGQLIADD278 pKa = 4.39LTDD281 pKa = 3.12ATAATVNQVRR291 pKa = 11.84LAFQTQKK298 pKa = 10.58FYY300 pKa = 11.35EE301 pKa = 4.17KK302 pKa = 9.84QARR305 pKa = 11.84GGTRR309 pKa = 11.84YY310 pKa = 8.49TEE312 pKa = 4.33VIKK315 pKa = 10.96SHH317 pKa = 6.92FDD319 pKa = 3.35VVSPDD324 pKa = 3.11MRR326 pKa = 11.84LQRR329 pKa = 11.84PEE331 pKa = 4.1YY332 pKa = 9.65IGGGSTPVMINPVAQTAPTSGSNALAQLGAYY363 pKa = 7.44GVCAPEE369 pKa = 3.85AHH371 pKa = 6.5GFTYY375 pKa = 10.53SATEE379 pKa = 3.67HH380 pKa = 5.9GVIVGLINVRR390 pKa = 11.84ADD392 pKa = 3.43LNYY395 pKa = 10.4QSGLDD400 pKa = 3.81RR401 pKa = 11.84MWTRR405 pKa = 11.84QVFTDD410 pKa = 3.84YY411 pKa = 9.81YY412 pKa = 9.27WPVFSHH418 pKa = 6.89LGEE421 pKa = 3.99QAVLNQEE428 pKa = 4.66IYY430 pKa = 10.92AQGTSADD437 pKa = 3.65TDD439 pKa = 3.54VFGYY443 pKa = 7.41QEE445 pKa = 3.49RR446 pKa = 11.84HH447 pKa = 4.66AEE449 pKa = 3.95YY450 pKa = 9.88RR451 pKa = 11.84YY452 pKa = 10.22KK453 pKa = 10.72PSLITGLFRR462 pKa = 11.84SNATGTLDD470 pKa = 3.44SWHH473 pKa = 6.89LAQDD477 pKa = 4.29FGSLPEE483 pKa = 4.89LNDD486 pKa = 3.27EE487 pKa = 5.46FIVEE491 pKa = 4.52DD492 pKa = 4.1APMSRR497 pKa = 11.84IKK499 pKa = 10.7AVASSYY505 pKa = 11.59DD506 pKa = 3.43FLFDD510 pKa = 4.12AVFAYY515 pKa = 10.2HH516 pKa = 7.24CARR519 pKa = 11.84PMPVYY524 pKa = 10.31AVPGLIDD531 pKa = 3.33HH532 pKa = 7.15FF533 pKa = 4.9
Molecular weight: 58.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W648|A0A5Q2W648_9VIRU Uncharacterized protein OS=Microviridae sp. ctnrr37 OX=2656704 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.31LWLRR6 pKa = 11.84QNRR9 pKa = 11.84ILNLRR14 pKa = 11.84HH15 pKa = 6.52NILINNLNLILRR27 pKa = 11.84HH28 pKa = 5.77NYY30 pKa = 9.07INNHH34 pKa = 5.33SLSIFRR40 pKa = 11.84IKK42 pKa = 10.05TLVRR46 pKa = 11.84ASKK49 pKa = 10.24HH50 pKa = 4.56FKK52 pKa = 10.26RR53 pKa = 11.84VAKK56 pKa = 9.78RR57 pKa = 11.84LKK59 pKa = 10.4VEE61 pKa = 4.37LSAISSPSSRR71 pKa = 11.84RR72 pKa = 11.84DD73 pKa = 2.92SFAFAKK79 pKa = 10.34PSGSSRR85 pKa = 11.84SHH87 pKa = 6.85KK88 pKa = 10.4ISS90 pKa = 3.02
MM1 pKa = 7.51KK2 pKa = 10.31LWLRR6 pKa = 11.84QNRR9 pKa = 11.84ILNLRR14 pKa = 11.84HH15 pKa = 6.52NILINNLNLILRR27 pKa = 11.84HH28 pKa = 5.77NYY30 pKa = 9.07INNHH34 pKa = 5.33SLSIFRR40 pKa = 11.84IKK42 pKa = 10.05TLVRR46 pKa = 11.84ASKK49 pKa = 10.24HH50 pKa = 4.56FKK52 pKa = 10.26RR53 pKa = 11.84VAKK56 pKa = 9.78RR57 pKa = 11.84LKK59 pKa = 10.4VEE61 pKa = 4.37LSAISSPSSRR71 pKa = 11.84RR72 pKa = 11.84DD73 pKa = 2.92SFAFAKK79 pKa = 10.34PSGSSRR85 pKa = 11.84SHH87 pKa = 6.85KK88 pKa = 10.4ISS90 pKa = 3.02
Molecular weight: 10.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1145 |
87 |
533 |
190.8 |
21.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.48 ± 1.96 | 1.048 ± 0.455 |
5.59 ± 0.762 | 4.017 ± 0.382 |
5.939 ± 0.492 | 8.384 ± 1.438 |
1.747 ± 0.567 | 3.93 ± 0.845 |
4.454 ± 1.457 | 7.773 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.231 ± 0.701 | 4.367 ± 0.681 |
5.153 ± 1.015 | 5.066 ± 1.016 |
6.114 ± 1.259 | 7.511 ± 1.187 |
5.939 ± 1.461 | 4.978 ± 0.71 |
1.223 ± 0.206 | 3.057 ± 0.795 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
