
Neodiprion lecontei nucleopolyhedrovirus (strain Canada) (NeleNPV)
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Baculoviridae; Gammabaculovirus; Neodiprion lecontei nucleopolyhedrovirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
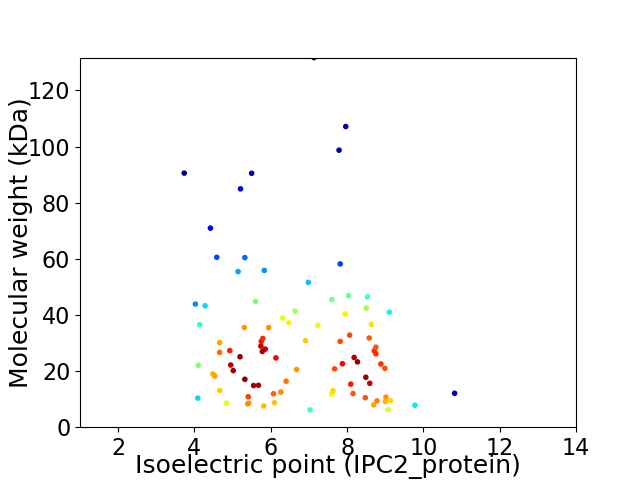
Virtual 2D-PAGE plot for 89 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6JP86|Q6JP86_NPVNC Uncharacterized protein OS=Neodiprion lecontei nucleopolyhedrovirus (strain Canada) OX=654906 PE=4 SV=1
MM1 pKa = 7.18FLIVFLFLCILLLFFTYY18 pKa = 10.95NFFVDD23 pKa = 3.24NTDD26 pKa = 3.03NVEE29 pKa = 4.11KK30 pKa = 10.74YY31 pKa = 10.2KK32 pKa = 10.65QIHH35 pKa = 5.14XLLMEE40 pKa = 5.84AYY42 pKa = 8.67TSDD45 pKa = 3.74IISFDD50 pKa = 3.28NVSFISRR57 pKa = 11.84VRR59 pKa = 11.84WPVYY63 pKa = 9.66EE64 pKa = 3.99VVTFEE69 pKa = 4.31TEE71 pKa = 4.31TYY73 pKa = 10.89TMTSNMWYY81 pKa = 10.15SGLEE85 pKa = 3.76KK86 pKa = 10.49RR87 pKa = 11.84FNFYY91 pKa = 11.01DD92 pKa = 3.38QTFEE96 pKa = 5.16DD97 pKa = 3.82VDD99 pKa = 4.44DD100 pKa = 3.83NTEE103 pKa = 3.88RR104 pKa = 11.84VTTIDD109 pKa = 3.09ATSFSLHH116 pKa = 6.7VDD118 pKa = 3.8DD119 pKa = 5.72GWVDD123 pKa = 4.3LKK125 pKa = 11.34CPANMIFDD133 pKa = 5.21GYY135 pKa = 10.73QCVRR139 pKa = 11.84QNICEE144 pKa = 4.04NQSAGTILPVNEE156 pKa = 4.31SLIDD160 pKa = 3.11IAFYY164 pKa = 10.69QKK166 pKa = 10.9YY167 pKa = 9.73NRR169 pKa = 11.84STTTDD174 pKa = 3.78TIYY177 pKa = 10.88HH178 pKa = 5.49PTLYY182 pKa = 9.83IVCNGDD188 pKa = 3.13DD189 pKa = 3.44SYY191 pKa = 11.72EE192 pKa = 4.15IKK194 pKa = 10.77EE195 pKa = 4.38CDD197 pKa = 3.6DD198 pKa = 3.63NQLFDD203 pKa = 5.89SDD205 pKa = 4.13MLQCVDD211 pKa = 4.76KK212 pKa = 10.96DD213 pKa = 3.45ACADD217 pKa = 3.49VADD220 pKa = 4.28YY221 pKa = 10.35FVLSDD226 pKa = 4.96RR227 pKa = 11.84PDD229 pKa = 3.29TLADD233 pKa = 4.4DD234 pKa = 4.76EE235 pKa = 5.75YY236 pKa = 11.89LMCLNNEE243 pKa = 4.37TTVQTCDD250 pKa = 2.75EE251 pKa = 4.16NMIFDD256 pKa = 4.36TTTLGCVEE264 pKa = 4.36VDD266 pKa = 3.11LCKK269 pKa = 10.5INGSGYY275 pKa = 9.34TYY277 pKa = 9.61ITDD280 pKa = 4.14DD281 pKa = 4.26LADD284 pKa = 3.56NQYY287 pKa = 9.73YY288 pKa = 9.68QCTSAFDD295 pKa = 3.77KK296 pKa = 11.14QIISCPIRR304 pKa = 11.84VEE306 pKa = 3.72TDD308 pKa = 3.01GQYY311 pKa = 11.11SCAGDD316 pKa = 3.85TRR318 pKa = 11.84CMSLEE323 pKa = 4.17NGSGSIMTMIEE334 pKa = 3.95NTHH337 pKa = 5.44FRR339 pKa = 11.84YY340 pKa = 10.48ANEE343 pKa = 4.01AVVCTDD349 pKa = 3.24YY350 pKa = 11.33EE351 pKa = 4.14ITDD354 pKa = 3.79SVVCDD359 pKa = 3.61SEE361 pKa = 5.23EE362 pKa = 3.94IEE364 pKa = 4.31LTVEE368 pKa = 4.47PFTLDD373 pKa = 3.19LXIPQIILDD382 pKa = 4.0RR383 pKa = 11.84TTNICKK389 pKa = 10.41SFDD392 pKa = 3.54INDD395 pKa = 3.33VEE397 pKa = 5.44IITDD401 pKa = 3.39HH402 pKa = 6.63AGIVSTEE409 pKa = 3.23NDD411 pKa = 3.55YY412 pKa = 11.7NVDD415 pKa = 3.41FTTAVRR421 pKa = 11.84VNVQNLLSKK430 pKa = 10.15NLPVSTSMLFSSLDD444 pKa = 3.32YY445 pKa = 10.88AKK447 pKa = 10.67NYY449 pKa = 10.01GFIGFSPLTGNEE461 pKa = 3.6IDD463 pKa = 4.18CYY465 pKa = 11.39GXVLYY470 pKa = 10.58DD471 pKa = 3.43IFAGDD476 pKa = 4.17KK477 pKa = 11.25YY478 pKa = 11.08NICSDD483 pKa = 3.96NEE485 pKa = 4.13ITSSNVIXDD494 pKa = 4.02NEE496 pKa = 4.12FFSTYY501 pKa = 9.26TNTVGTTSITSPCRR515 pKa = 11.84VKK517 pKa = 11.03SDD519 pKa = 2.99IVLDD523 pKa = 3.5NDD525 pKa = 3.97IYY527 pKa = 10.83IQEE530 pKa = 4.15SDD532 pKa = 3.74NVSVSCLYY540 pKa = 11.18AMPLQEE546 pKa = 6.1IIVTDD551 pKa = 4.09LAEE554 pKa = 4.25TAGVLVNDD562 pKa = 3.74EE563 pKa = 4.33CFEE566 pKa = 4.72NGIDD570 pKa = 3.86LNNVAGAIVILNDD583 pKa = 3.2EE584 pKa = 4.31LACRR588 pKa = 11.84SVYY591 pKa = 10.36VEE593 pKa = 4.53SYY595 pKa = 9.05NTIITASFTPVMKK608 pKa = 10.69FEE610 pKa = 3.99EE611 pKa = 4.54RR612 pKa = 11.84TWNVDD617 pKa = 3.12TQSYY621 pKa = 10.09DD622 pKa = 2.77GVMYY626 pKa = 10.6DD627 pKa = 3.81SVLHH631 pKa = 5.73KK632 pKa = 7.79TTNGWLACPPSLYY645 pKa = 10.73DD646 pKa = 3.97EE647 pKa = 4.42TTMDD651 pKa = 4.56CNIDD655 pKa = 3.33KK656 pKa = 10.24TKK658 pKa = 10.85LYY660 pKa = 9.73TIRR663 pKa = 11.84DD664 pKa = 3.44LHH666 pKa = 6.28FVQNYY671 pKa = 9.38PDD673 pKa = 3.74IDD675 pKa = 3.94LEE677 pKa = 4.4AEE679 pKa = 3.9NEE681 pKa = 4.11INIDD685 pKa = 3.45TDD687 pKa = 4.19DD688 pKa = 4.11VDD690 pKa = 3.75VSNPIVDD697 pKa = 4.58EE698 pKa = 4.24IEE700 pKa = 4.68AINPIVDD707 pKa = 3.61DD708 pKa = 3.93VVLRR712 pKa = 11.84RR713 pKa = 11.84INNATNNIHH722 pKa = 7.44PILEE726 pKa = 4.26NKK728 pKa = 9.07VVSDD732 pKa = 3.93STQMTLDD739 pKa = 3.47KK740 pKa = 10.57KK741 pKa = 10.7EE742 pKa = 4.1PSIDD746 pKa = 3.61VQDD749 pKa = 3.79IDD751 pKa = 4.06STSTSSNEE759 pKa = 3.83SAEE762 pKa = 4.21NEE764 pKa = 4.43STVLDD769 pKa = 3.92PSKK772 pKa = 10.59VLEE775 pKa = 4.4YY776 pKa = 10.34INRR779 pKa = 11.84YY780 pKa = 8.98HH781 pKa = 7.41EE782 pKa = 4.69SDD784 pKa = 3.08TSEE787 pKa = 4.08EE788 pKa = 4.04EE789 pKa = 4.4EE790 pKa = 4.52YY791 pKa = 10.55MDD793 pKa = 6.01DD794 pKa = 4.55SDD796 pKa = 4.37ILASLFEE803 pKa = 4.25
MM1 pKa = 7.18FLIVFLFLCILLLFFTYY18 pKa = 10.95NFFVDD23 pKa = 3.24NTDD26 pKa = 3.03NVEE29 pKa = 4.11KK30 pKa = 10.74YY31 pKa = 10.2KK32 pKa = 10.65QIHH35 pKa = 5.14XLLMEE40 pKa = 5.84AYY42 pKa = 8.67TSDD45 pKa = 3.74IISFDD50 pKa = 3.28NVSFISRR57 pKa = 11.84VRR59 pKa = 11.84WPVYY63 pKa = 9.66EE64 pKa = 3.99VVTFEE69 pKa = 4.31TEE71 pKa = 4.31TYY73 pKa = 10.89TMTSNMWYY81 pKa = 10.15SGLEE85 pKa = 3.76KK86 pKa = 10.49RR87 pKa = 11.84FNFYY91 pKa = 11.01DD92 pKa = 3.38QTFEE96 pKa = 5.16DD97 pKa = 3.82VDD99 pKa = 4.44DD100 pKa = 3.83NTEE103 pKa = 3.88RR104 pKa = 11.84VTTIDD109 pKa = 3.09ATSFSLHH116 pKa = 6.7VDD118 pKa = 3.8DD119 pKa = 5.72GWVDD123 pKa = 4.3LKK125 pKa = 11.34CPANMIFDD133 pKa = 5.21GYY135 pKa = 10.73QCVRR139 pKa = 11.84QNICEE144 pKa = 4.04NQSAGTILPVNEE156 pKa = 4.31SLIDD160 pKa = 3.11IAFYY164 pKa = 10.69QKK166 pKa = 10.9YY167 pKa = 9.73NRR169 pKa = 11.84STTTDD174 pKa = 3.78TIYY177 pKa = 10.88HH178 pKa = 5.49PTLYY182 pKa = 9.83IVCNGDD188 pKa = 3.13DD189 pKa = 3.44SYY191 pKa = 11.72EE192 pKa = 4.15IKK194 pKa = 10.77EE195 pKa = 4.38CDD197 pKa = 3.6DD198 pKa = 3.63NQLFDD203 pKa = 5.89SDD205 pKa = 4.13MLQCVDD211 pKa = 4.76KK212 pKa = 10.96DD213 pKa = 3.45ACADD217 pKa = 3.49VADD220 pKa = 4.28YY221 pKa = 10.35FVLSDD226 pKa = 4.96RR227 pKa = 11.84PDD229 pKa = 3.29TLADD233 pKa = 4.4DD234 pKa = 4.76EE235 pKa = 5.75YY236 pKa = 11.89LMCLNNEE243 pKa = 4.37TTVQTCDD250 pKa = 2.75EE251 pKa = 4.16NMIFDD256 pKa = 4.36TTTLGCVEE264 pKa = 4.36VDD266 pKa = 3.11LCKK269 pKa = 10.5INGSGYY275 pKa = 9.34TYY277 pKa = 9.61ITDD280 pKa = 4.14DD281 pKa = 4.26LADD284 pKa = 3.56NQYY287 pKa = 9.73YY288 pKa = 9.68QCTSAFDD295 pKa = 3.77KK296 pKa = 11.14QIISCPIRR304 pKa = 11.84VEE306 pKa = 3.72TDD308 pKa = 3.01GQYY311 pKa = 11.11SCAGDD316 pKa = 3.85TRR318 pKa = 11.84CMSLEE323 pKa = 4.17NGSGSIMTMIEE334 pKa = 3.95NTHH337 pKa = 5.44FRR339 pKa = 11.84YY340 pKa = 10.48ANEE343 pKa = 4.01AVVCTDD349 pKa = 3.24YY350 pKa = 11.33EE351 pKa = 4.14ITDD354 pKa = 3.79SVVCDD359 pKa = 3.61SEE361 pKa = 5.23EE362 pKa = 3.94IEE364 pKa = 4.31LTVEE368 pKa = 4.47PFTLDD373 pKa = 3.19LXIPQIILDD382 pKa = 4.0RR383 pKa = 11.84TTNICKK389 pKa = 10.41SFDD392 pKa = 3.54INDD395 pKa = 3.33VEE397 pKa = 5.44IITDD401 pKa = 3.39HH402 pKa = 6.63AGIVSTEE409 pKa = 3.23NDD411 pKa = 3.55YY412 pKa = 11.7NVDD415 pKa = 3.41FTTAVRR421 pKa = 11.84VNVQNLLSKK430 pKa = 10.15NLPVSTSMLFSSLDD444 pKa = 3.32YY445 pKa = 10.88AKK447 pKa = 10.67NYY449 pKa = 10.01GFIGFSPLTGNEE461 pKa = 3.6IDD463 pKa = 4.18CYY465 pKa = 11.39GXVLYY470 pKa = 10.58DD471 pKa = 3.43IFAGDD476 pKa = 4.17KK477 pKa = 11.25YY478 pKa = 11.08NICSDD483 pKa = 3.96NEE485 pKa = 4.13ITSSNVIXDD494 pKa = 4.02NEE496 pKa = 4.12FFSTYY501 pKa = 9.26TNTVGTTSITSPCRR515 pKa = 11.84VKK517 pKa = 11.03SDD519 pKa = 2.99IVLDD523 pKa = 3.5NDD525 pKa = 3.97IYY527 pKa = 10.83IQEE530 pKa = 4.15SDD532 pKa = 3.74NVSVSCLYY540 pKa = 11.18AMPLQEE546 pKa = 6.1IIVTDD551 pKa = 4.09LAEE554 pKa = 4.25TAGVLVNDD562 pKa = 3.74EE563 pKa = 4.33CFEE566 pKa = 4.72NGIDD570 pKa = 3.86LNNVAGAIVILNDD583 pKa = 3.2EE584 pKa = 4.31LACRR588 pKa = 11.84SVYY591 pKa = 10.36VEE593 pKa = 4.53SYY595 pKa = 9.05NTIITASFTPVMKK608 pKa = 10.69FEE610 pKa = 3.99EE611 pKa = 4.54RR612 pKa = 11.84TWNVDD617 pKa = 3.12TQSYY621 pKa = 10.09DD622 pKa = 2.77GVMYY626 pKa = 10.6DD627 pKa = 3.81SVLHH631 pKa = 5.73KK632 pKa = 7.79TTNGWLACPPSLYY645 pKa = 10.73DD646 pKa = 3.97EE647 pKa = 4.42TTMDD651 pKa = 4.56CNIDD655 pKa = 3.33KK656 pKa = 10.24TKK658 pKa = 10.85LYY660 pKa = 9.73TIRR663 pKa = 11.84DD664 pKa = 3.44LHH666 pKa = 6.28FVQNYY671 pKa = 9.38PDD673 pKa = 3.74IDD675 pKa = 3.94LEE677 pKa = 4.4AEE679 pKa = 3.9NEE681 pKa = 4.11INIDD685 pKa = 3.45TDD687 pKa = 4.19DD688 pKa = 4.11VDD690 pKa = 3.75VSNPIVDD697 pKa = 4.58EE698 pKa = 4.24IEE700 pKa = 4.68AINPIVDD707 pKa = 3.61DD708 pKa = 3.93VVLRR712 pKa = 11.84RR713 pKa = 11.84INNATNNIHH722 pKa = 7.44PILEE726 pKa = 4.26NKK728 pKa = 9.07VVSDD732 pKa = 3.93STQMTLDD739 pKa = 3.47KK740 pKa = 10.57KK741 pKa = 10.7EE742 pKa = 4.1PSIDD746 pKa = 3.61VQDD749 pKa = 3.79IDD751 pKa = 4.06STSTSSNEE759 pKa = 3.83SAEE762 pKa = 4.21NEE764 pKa = 4.43STVLDD769 pKa = 3.92PSKK772 pKa = 10.59VLEE775 pKa = 4.4YY776 pKa = 10.34INRR779 pKa = 11.84YY780 pKa = 8.98HH781 pKa = 7.41EE782 pKa = 4.69SDD784 pKa = 3.08TSEE787 pKa = 4.08EE788 pKa = 4.04EE789 pKa = 4.4EE790 pKa = 4.52YY791 pKa = 10.55MDD793 pKa = 6.01DD794 pKa = 4.55SDD796 pKa = 4.37ILASLFEE803 pKa = 4.25
Molecular weight: 90.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6JPE2|Q6JPE2_NPVNC Uncharacterized protein OS=Neodiprion lecontei nucleopolyhedrovirus (strain Canada) OX=654906 PE=4 SV=1
MM1 pKa = 7.92VSEE4 pKa = 4.59SSDD7 pKa = 3.28SKK9 pKa = 11.53KK10 pKa = 10.0EE11 pKa = 3.91LKK13 pKa = 10.16PIEE16 pKa = 3.71IMAYY20 pKa = 8.29NRR22 pKa = 11.84RR23 pKa = 11.84PGRR26 pKa = 11.84PRR28 pKa = 11.84KK29 pKa = 9.48RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 11.43GNNSTNRR40 pKa = 11.84TRR42 pKa = 11.84SRR44 pKa = 11.84SRR46 pKa = 11.84SRR48 pKa = 11.84SRR50 pKa = 11.84SRR52 pKa = 11.84SRR54 pKa = 11.84SRR56 pKa = 11.84SRR58 pKa = 11.84TGGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84SRR66 pKa = 11.84SSSSYY71 pKa = 8.19NRR73 pKa = 11.84NYY75 pKa = 10.37RR76 pKa = 11.84SSNSPSSNRR85 pKa = 11.84QYY87 pKa = 11.02GRR89 pKa = 11.84SRR91 pKa = 11.84SRR93 pKa = 11.84SRR95 pKa = 11.84SLSQRR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 9.37
MM1 pKa = 7.92VSEE4 pKa = 4.59SSDD7 pKa = 3.28SKK9 pKa = 11.53KK10 pKa = 10.0EE11 pKa = 3.91LKK13 pKa = 10.16PIEE16 pKa = 3.71IMAYY20 pKa = 8.29NRR22 pKa = 11.84RR23 pKa = 11.84PGRR26 pKa = 11.84PRR28 pKa = 11.84KK29 pKa = 9.48RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 11.43GNNSTNRR40 pKa = 11.84TRR42 pKa = 11.84SRR44 pKa = 11.84SRR46 pKa = 11.84SRR48 pKa = 11.84SRR50 pKa = 11.84SRR52 pKa = 11.84SRR54 pKa = 11.84SRR56 pKa = 11.84SRR58 pKa = 11.84TGGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84SRR66 pKa = 11.84SSSSYY71 pKa = 8.19NRR73 pKa = 11.84NYY75 pKa = 10.37RR76 pKa = 11.84SSNSPSSNRR85 pKa = 11.84QYY87 pKa = 11.02GRR89 pKa = 11.84SRR91 pKa = 11.84SRR93 pKa = 11.84SRR95 pKa = 11.84SLSQRR100 pKa = 11.84RR101 pKa = 11.84YY102 pKa = 9.37
Molecular weight: 12.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
24124 |
52 |
1134 |
271.1 |
31.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.465 ± 0.206 | 2.636 ± 0.163 |
6.587 ± 0.208 | 5.186 ± 0.2 |
5.007 ± 0.204 | 2.922 ± 0.198 |
2.413 ± 0.152 | 8.983 ± 0.187 |
7.184 ± 0.299 | 8.966 ± 0.194 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.317 ± 0.093 | 8.079 ± 0.233 |
2.657 ± 0.133 | 3.457 ± 0.128 |
4.149 ± 0.217 | 6.997 ± 0.22 |
6.848 ± 0.23 | 6.334 ± 0.164 |
0.767 ± 0.071 | 4.825 ± 0.149 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
