
Diplodia seriata
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetes incertae sedis; Botryosphaeriales; Botryosphaeriaceae; Diplodia
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
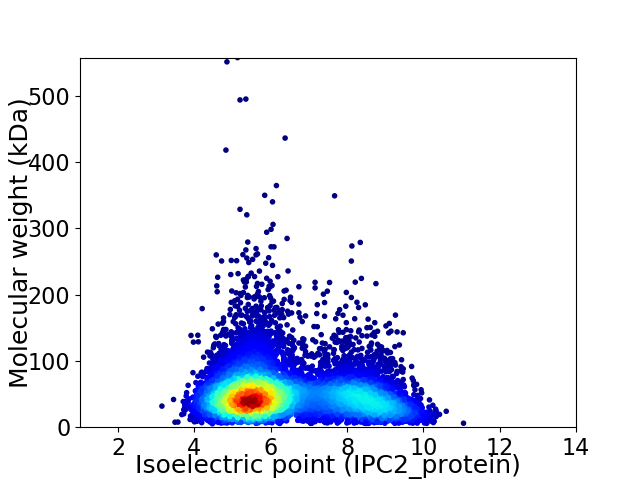
Virtual 2D-PAGE plot for 8038 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
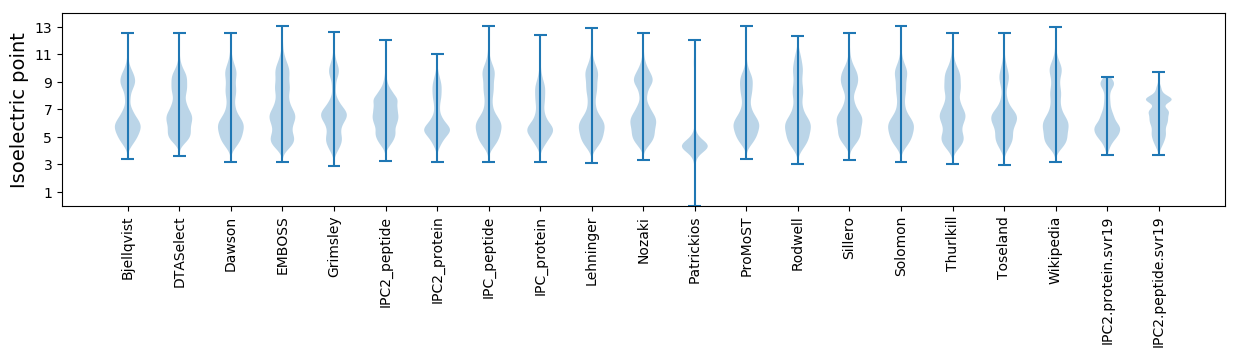
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S8B9C2|A0A1S8B9C2_9PEZI ATP-grasp domain-containing protein OS=Diplodia seriata OX=420778 GN=BK809_0001532 PE=4 SV=1
MM1 pKa = 6.88VQINNALAGMIVGAGALVAAAPAPAPTAAPNLKK34 pKa = 9.89DD35 pKa = 3.43AQIGKK40 pKa = 9.59RR41 pKa = 11.84ADD43 pKa = 2.99ACTFTDD49 pKa = 4.24ASAASASKK57 pKa = 10.46ASCSTIILSDD67 pKa = 3.02ITVPAGEE74 pKa = 4.4TLDD77 pKa = 3.72MTDD80 pKa = 5.37LEE82 pKa = 5.02DD83 pKa = 3.37NTYY86 pKa = 10.71VEE88 pKa = 4.74FQGTTTFEE96 pKa = 3.9YY97 pKa = 10.94DD98 pKa = 2.94EE99 pKa = 4.51WEE101 pKa = 4.63GPLVSFSGTGITITGASGHH120 pKa = 5.71VLDD123 pKa = 5.83GNGAKK128 pKa = 8.87WWDD131 pKa = 3.73GEE133 pKa = 4.25GSNGGVTKK141 pKa = 10.54PKK143 pKa = 10.33FFYY146 pKa = 10.52AHH148 pKa = 6.06SLKK151 pKa = 10.9SSTIKK156 pKa = 10.71GLNIKK161 pKa = 7.66NTPVQCMSINGAEE174 pKa = 4.11EE175 pKa = 6.03LYY177 pKa = 11.04VQDD180 pKa = 3.45ITIDD184 pKa = 3.71NTDD187 pKa = 3.35GDD189 pKa = 4.27ATNDD193 pKa = 3.55DD194 pKa = 4.3GDD196 pKa = 4.35ALGHH200 pKa = 5.75NTDD203 pKa = 3.95AFDD206 pKa = 3.84IGSSTGVYY214 pKa = 9.77ISGANVQNQDD224 pKa = 2.67DD225 pKa = 4.17CMAINSGEE233 pKa = 4.49NISMMDD239 pKa = 3.81GTCSGGHH246 pKa = 5.92GLSIGSVGGRR256 pKa = 11.84SDD258 pKa = 4.44NDD260 pKa = 3.39VKK262 pKa = 11.25NVTVSGMTVKK272 pKa = 10.67NSQNGVRR279 pKa = 11.84IKK281 pKa = 9.73TVYY284 pKa = 10.35DD285 pKa = 3.33ATGSVSDD292 pKa = 3.93ITYY295 pKa = 10.91SDD297 pKa = 3.18ITLSGITKK305 pKa = 8.93YY306 pKa = 10.97GIVIEE311 pKa = 3.95QDD313 pKa = 3.68YY314 pKa = 11.2EE315 pKa = 4.22NGSPTGEE322 pKa = 4.18PTDD325 pKa = 3.87GVPITGLTIDD335 pKa = 4.09GVTGSVEE342 pKa = 4.5DD343 pKa = 4.62DD344 pKa = 3.19ATPIYY349 pKa = 10.05ILCADD354 pKa = 4.99GACSDD359 pKa = 4.07WSWSADD365 pKa = 3.1ITGGEE370 pKa = 4.19ASTEE374 pKa = 4.0CSGIPDD380 pKa = 4.02GASCC384 pKa = 5.2
MM1 pKa = 6.88VQINNALAGMIVGAGALVAAAPAPAPTAAPNLKK34 pKa = 9.89DD35 pKa = 3.43AQIGKK40 pKa = 9.59RR41 pKa = 11.84ADD43 pKa = 2.99ACTFTDD49 pKa = 4.24ASAASASKK57 pKa = 10.46ASCSTIILSDD67 pKa = 3.02ITVPAGEE74 pKa = 4.4TLDD77 pKa = 3.72MTDD80 pKa = 5.37LEE82 pKa = 5.02DD83 pKa = 3.37NTYY86 pKa = 10.71VEE88 pKa = 4.74FQGTTTFEE96 pKa = 3.9YY97 pKa = 10.94DD98 pKa = 2.94EE99 pKa = 4.51WEE101 pKa = 4.63GPLVSFSGTGITITGASGHH120 pKa = 5.71VLDD123 pKa = 5.83GNGAKK128 pKa = 8.87WWDD131 pKa = 3.73GEE133 pKa = 4.25GSNGGVTKK141 pKa = 10.54PKK143 pKa = 10.33FFYY146 pKa = 10.52AHH148 pKa = 6.06SLKK151 pKa = 10.9SSTIKK156 pKa = 10.71GLNIKK161 pKa = 7.66NTPVQCMSINGAEE174 pKa = 4.11EE175 pKa = 6.03LYY177 pKa = 11.04VQDD180 pKa = 3.45ITIDD184 pKa = 3.71NTDD187 pKa = 3.35GDD189 pKa = 4.27ATNDD193 pKa = 3.55DD194 pKa = 4.3GDD196 pKa = 4.35ALGHH200 pKa = 5.75NTDD203 pKa = 3.95AFDD206 pKa = 3.84IGSSTGVYY214 pKa = 9.77ISGANVQNQDD224 pKa = 2.67DD225 pKa = 4.17CMAINSGEE233 pKa = 4.49NISMMDD239 pKa = 3.81GTCSGGHH246 pKa = 5.92GLSIGSVGGRR256 pKa = 11.84SDD258 pKa = 4.44NDD260 pKa = 3.39VKK262 pKa = 11.25NVTVSGMTVKK272 pKa = 10.67NSQNGVRR279 pKa = 11.84IKK281 pKa = 9.73TVYY284 pKa = 10.35DD285 pKa = 3.33ATGSVSDD292 pKa = 3.93ITYY295 pKa = 10.91SDD297 pKa = 3.18ITLSGITKK305 pKa = 8.93YY306 pKa = 10.97GIVIEE311 pKa = 3.95QDD313 pKa = 3.68YY314 pKa = 11.2EE315 pKa = 4.22NGSPTGEE322 pKa = 4.18PTDD325 pKa = 3.87GVPITGLTIDD335 pKa = 4.09GVTGSVEE342 pKa = 4.5DD343 pKa = 4.62DD344 pKa = 3.19ATPIYY349 pKa = 10.05ILCADD354 pKa = 4.99GACSDD359 pKa = 4.07WSWSADD365 pKa = 3.1ITGGEE370 pKa = 4.19ASTEE374 pKa = 4.0CSGIPDD380 pKa = 4.02GASCC384 pKa = 5.2
Molecular weight: 39.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S8BP68|A0A1S8BP68_9PEZI MBOAT_2 domain-containing protein OS=Diplodia seriata OX=420778 GN=BK809_0005957 PE=4 SV=1
SS1 pKa = 6.27QKK3 pKa = 10.7SFRR6 pKa = 11.84TKK8 pKa = 10.38QKK10 pKa = 9.84LAKK13 pKa = 9.55AQKK16 pKa = 8.59QNRR19 pKa = 11.84PIPQWIRR26 pKa = 11.84LRR28 pKa = 11.84TGNTIRR34 pKa = 11.84YY35 pKa = 5.79NAKK38 pKa = 8.89RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 4.15WRR43 pKa = 11.84KK44 pKa = 7.38TRR46 pKa = 11.84IGII49 pKa = 4.0
SS1 pKa = 6.27QKK3 pKa = 10.7SFRR6 pKa = 11.84TKK8 pKa = 10.38QKK10 pKa = 9.84LAKK13 pKa = 9.55AQKK16 pKa = 8.59QNRR19 pKa = 11.84PIPQWIRR26 pKa = 11.84LRR28 pKa = 11.84TGNTIRR34 pKa = 11.84YY35 pKa = 5.79NAKK38 pKa = 8.89RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 4.15WRR43 pKa = 11.84KK44 pKa = 7.38TRR46 pKa = 11.84IGII49 pKa = 4.0
Molecular weight: 6.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4002348 |
49 |
5079 |
497.9 |
54.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.719 ± 0.029 | 1.141 ± 0.009 |
5.828 ± 0.018 | 6.244 ± 0.028 |
3.681 ± 0.016 | 7.414 ± 0.029 |
2.349 ± 0.011 | 4.351 ± 0.017 |
4.74 ± 0.026 | 8.666 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.129 ± 0.01 | 3.474 ± 0.013 |
6.101 ± 0.027 | 3.863 ± 0.02 |
6.24 ± 0.023 | 7.744 ± 0.034 |
5.759 ± 0.017 | 6.357 ± 0.021 |
1.489 ± 0.011 | 2.712 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
