
Trinickia symbiotica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Trinickia
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
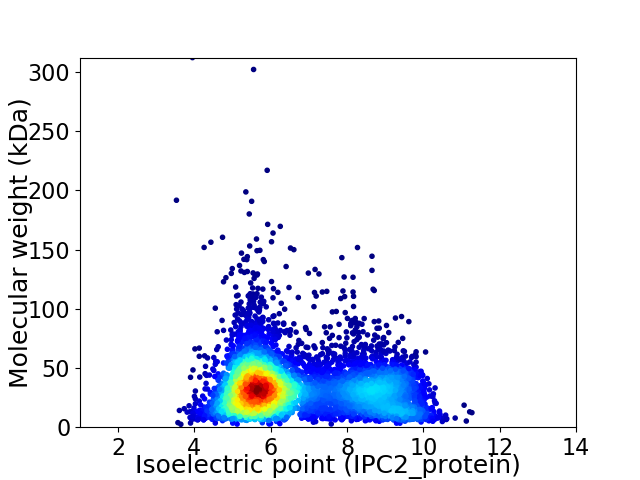
Virtual 2D-PAGE plot for 5982 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N7WKB9|A0A2N7WKB9_9BURK MBL fold metallo-hydrolase OS=Trinickia symbiotica OX=863227 GN=C0Z20_30580 PE=4 SV=1
MM1 pKa = 8.08DD2 pKa = 4.31AQEE5 pKa = 4.8LGVLEE10 pKa = 4.87KK11 pKa = 10.28FTGEE15 pKa = 3.94NFVSSFTGGPDD26 pKa = 3.22PSNWHH31 pKa = 5.16QAQVLQDD38 pKa = 3.79AFNQLMNAVEE48 pKa = 4.27VRR50 pKa = 11.84LVAQGPLASYY60 pKa = 10.84LSGVGFNYY68 pKa = 9.13DD69 pKa = 3.04TDD71 pKa = 4.22SLVGSVDD78 pKa = 3.5LTALAEE84 pKa = 4.12ALINATPAGQLAAEE98 pKa = 5.45GYY100 pKa = 7.67WSSVAPMVGNLCSEE114 pKa = 4.82LGLGASAYY122 pKa = 7.54QTAFQGPFSSLNLPFSLDD140 pKa = 3.67EE141 pKa = 4.0IVQGNVFLGNGTNTVAVAHH160 pKa = 5.83TAGVPHH166 pKa = 6.65YY167 pKa = 9.9FDD169 pKa = 4.47GGPGVRR175 pKa = 11.84YY176 pKa = 9.36EE177 pKa = 4.27GSSGSGDD184 pKa = 3.02VFVFNQGYY192 pKa = 8.2GQLEE196 pKa = 4.05INEE199 pKa = 4.38FDD201 pKa = 3.37WNASNNVLRR210 pKa = 11.84LGAGITPDD218 pKa = 3.43AVKK221 pKa = 9.58VTLDD225 pKa = 3.73DD226 pKa = 4.0NQDD229 pKa = 3.01ILLTIGSNGDD239 pKa = 3.56QIKK242 pKa = 10.61LDD244 pKa = 3.85SEE246 pKa = 4.75GTGDD250 pKa = 4.02LRR252 pKa = 11.84WGVEE256 pKa = 3.89QVQFADD262 pKa = 4.13GAAWTQSQLIASARR276 pKa = 11.84NIQGTAASEE285 pKa = 4.05TLYY288 pKa = 9.62GTNGADD294 pKa = 3.2TFDD297 pKa = 4.52GKK299 pKa = 11.36GSTPYY304 pKa = 10.28SGSDD308 pKa = 3.36VEE310 pKa = 5.08IGNGGNDD317 pKa = 2.89TFIFNEE323 pKa = 5.21GYY325 pKa = 10.84GKK327 pKa = 8.74LTINEE332 pKa = 4.04YY333 pKa = 10.85DD334 pKa = 3.62YY335 pKa = 11.63GASPEE340 pKa = 4.15NVLQLGAGITPDD352 pKa = 3.32QVVVSVDD359 pKa = 3.04RR360 pKa = 11.84SQNIVLTIGTNGDD373 pKa = 3.05QVMLNRR379 pKa = 11.84EE380 pKa = 4.07ATGDD384 pKa = 3.39ARR386 pKa = 11.84EE387 pKa = 4.45GVQQIQFADD396 pKa = 3.94GTIWTQSQLLGMLATIDD413 pKa = 4.06GTPQDD418 pKa = 3.78DD419 pKa = 3.8TLYY422 pKa = 9.41GTGGANVFDD431 pKa = 4.41GKK433 pKa = 10.78GGYY436 pKa = 9.31DD437 pKa = 3.29VEE439 pKa = 4.63VGNGGNDD446 pKa = 3.13TYY448 pKa = 11.6VFNSGYY454 pKa = 8.15GTLVIKK460 pKa = 10.72DD461 pKa = 3.78SGSGVLDD468 pKa = 4.68LGTGITPSEE477 pKa = 4.19VKK479 pKa = 10.43ISTDD483 pKa = 3.25YY484 pKa = 11.55SGDD487 pKa = 3.21IVLTIGTEE495 pKa = 4.15GDD497 pKa = 3.42QVMLSGMTRR506 pKa = 11.84GASSSIQQVLFADD519 pKa = 4.59GTSWTAAQILAWSSDD534 pKa = 3.31IQGTTGNDD542 pKa = 3.33DD543 pKa = 4.12LYY545 pKa = 10.5GTSGDD550 pKa = 4.24DD551 pKa = 3.49VFDD554 pKa = 4.25GKK556 pKa = 11.08GGNDD560 pKa = 3.49YY561 pKa = 11.3EE562 pKa = 4.6SGRR565 pKa = 11.84GGNDD569 pKa = 2.83TFIFNQGYY577 pKa = 7.93GHH579 pKa = 7.37LEE581 pKa = 3.44ISEE584 pKa = 4.5VNGSAGFDD592 pKa = 3.41SVLEE596 pKa = 4.29LGSGITPSAVTIAYY610 pKa = 8.69GPNQSLVLTDD620 pKa = 4.41GNDD623 pKa = 3.24QITLDD628 pKa = 3.76GMANGSGRR636 pKa = 11.84DD637 pKa = 3.73GVQTVQFADD646 pKa = 3.9GTVWTASQMLASIGEE661 pKa = 4.06IRR663 pKa = 11.84GTAGDD668 pKa = 3.91DD669 pKa = 3.49QLFGTSGANLFDD681 pKa = 4.25GGGGNDD687 pKa = 3.82YY688 pKa = 11.36EE689 pKa = 6.34DD690 pKa = 4.83GDD692 pKa = 4.19GGNDD696 pKa = 3.06TFIFNRR702 pKa = 11.84GYY704 pKa = 8.6GTLEE708 pKa = 3.72IVEE711 pKa = 4.3QGAATTTGTLLLGPGILPSSVTVSLAGDD739 pKa = 3.88SVLLTIGSNGDD750 pKa = 3.66QVKK753 pKa = 10.4LDD755 pKa = 3.46YY756 pKa = 8.43MTDD759 pKa = 3.14RR760 pKa = 11.84GSNFGVRR767 pKa = 11.84DD768 pKa = 3.54VQFADD773 pKa = 3.73GTVWTASQILASLSDD788 pKa = 3.54VSGTAKK794 pKa = 10.36DD795 pKa = 3.8DD796 pKa = 4.62SIGGTSDD803 pKa = 3.27PSVIDD808 pKa = 3.75GKK810 pKa = 11.08GGNDD814 pKa = 3.14VAGGQGNNDD823 pKa = 3.27TFVFKK828 pKa = 10.72AGYY831 pKa = 9.4GHH833 pKa = 6.68LTVYY837 pKa = 10.11EE838 pKa = 4.03PQAANGVTNVLLFGPDD854 pKa = 2.87IDD856 pKa = 4.12EE857 pKa = 4.52SFVTVKK863 pKa = 10.91EE864 pKa = 4.01NMQDD868 pKa = 3.44AVVLTDD874 pKa = 5.28GIAGDD879 pKa = 4.13EE880 pKa = 4.23VTLVNQAYY888 pKa = 8.88EE889 pKa = 4.27FQGLQYY895 pKa = 11.13GGVQEE900 pKa = 4.31VQFADD905 pKa = 3.46GTMWTAQQLLQRR917 pKa = 11.84ATQGTPGDD925 pKa = 3.98DD926 pKa = 3.49VLIGGWQASLLDD938 pKa = 3.9GKK940 pKa = 10.9GGNDD944 pKa = 3.49YY945 pKa = 10.96EE946 pKa = 5.53AGGGDD951 pKa = 3.35GDD953 pKa = 3.83TFVFNAGYY961 pKa = 10.35GHH963 pKa = 6.7LQINEE968 pKa = 4.32YY969 pKa = 9.85QWGTANNVLQLGQGISEE986 pKa = 4.31SSISASVTRR995 pKa = 11.84GDD997 pKa = 5.25AIVLSDD1003 pKa = 4.41GTPGDD1008 pKa = 3.79QIVLDD1013 pKa = 4.44GEE1015 pKa = 4.33FDD1017 pKa = 3.56DD1018 pKa = 5.2ARR1020 pKa = 11.84YY1021 pKa = 10.21GGTSRR1026 pKa = 11.84GIQEE1030 pKa = 3.89VQFADD1035 pKa = 3.51GTTWSVQQLVQLAQTRR1051 pKa = 11.84TGTPGDD1057 pKa = 4.23DD1058 pKa = 3.62YY1059 pKa = 11.91LVGTTTASVFDD1070 pKa = 4.15GKK1072 pKa = 10.93GGNDD1076 pKa = 3.35FEE1078 pKa = 4.87TGGGKK1083 pKa = 10.5GDD1085 pKa = 3.32TFVFNAGYY1093 pKa = 9.82GHH1095 pKa = 7.08LEE1097 pKa = 3.9IYY1099 pKa = 9.18EE1100 pKa = 4.1YY1101 pKa = 9.92PYY1103 pKa = 10.97QPTNVLQLGPGISASSVEE1121 pKa = 4.29VSATEE1126 pKa = 4.08DD1127 pKa = 3.4YY1128 pKa = 11.41GVILTDD1134 pKa = 3.78GMAGDD1139 pKa = 4.71EE1140 pKa = 4.29IEE1142 pKa = 5.36LDD1144 pKa = 3.8GEE1146 pKa = 4.39LGGEE1150 pKa = 4.26GVEE1153 pKa = 4.01QVQFSDD1159 pKa = 3.48GTVWTRR1165 pKa = 11.84QQLLQMAEE1173 pKa = 4.39TTTGTPGDD1181 pKa = 4.16DD1182 pKa = 3.53VLLGTTDD1189 pKa = 3.41ASLFDD1194 pKa = 3.95GKK1196 pKa = 10.95GGNDD1200 pKa = 3.46YY1201 pKa = 11.6EE1202 pKa = 4.39EE1203 pKa = 5.86GDD1205 pKa = 4.26GNGHH1209 pKa = 7.5LDD1211 pKa = 3.29TFVFNAGYY1219 pKa = 9.93GHH1221 pKa = 7.63LEE1223 pKa = 3.8IDD1225 pKa = 3.46EE1226 pKa = 4.55SNYY1229 pKa = 9.7TVATSVLEE1237 pKa = 4.05LGPGISEE1244 pKa = 4.46SSLQVTTDD1252 pKa = 2.93KK1253 pKa = 11.64SEE1255 pKa = 5.36DD1256 pKa = 3.34IVLTSGTDD1264 pKa = 3.2QIVIDD1269 pKa = 4.21WMADD1273 pKa = 2.67GWGDD1277 pKa = 3.63YY1278 pKa = 11.07GVGQVTFADD1287 pKa = 4.17GTVWTASQIIAMASNVQGTAGNDD1310 pKa = 3.44TLYY1313 pKa = 9.57GTSISSTFDD1322 pKa = 3.02GKK1324 pKa = 11.04GGNDD1328 pKa = 3.29VEE1330 pKa = 4.66IGNGNDD1336 pKa = 3.12DD1337 pKa = 3.87TFIFDD1342 pKa = 3.78PGYY1345 pKa = 9.3GHH1347 pKa = 7.66LEE1349 pKa = 3.98IEE1351 pKa = 4.4EE1352 pKa = 4.28GMANYY1357 pKa = 10.64DD1358 pKa = 3.63STNVLQFGAGIRR1370 pKa = 11.84AAMVTASLGSNGSIVLTSGTDD1391 pKa = 3.43QVVLDD1396 pKa = 3.97GMTTQSCGIQLVQFADD1412 pKa = 4.14GTQWTPSQILAMASIIRR1429 pKa = 11.84GTAGNDD1435 pKa = 3.63TLSGTAGGDD1444 pKa = 3.05WFDD1447 pKa = 3.71GRR1449 pKa = 11.84GGNDD1453 pKa = 3.39LEE1455 pKa = 4.58VGGGGADD1462 pKa = 2.76TFVFNQGYY1470 pKa = 8.16GHH1472 pKa = 7.39LEE1474 pKa = 3.64IDD1476 pKa = 3.83EE1477 pKa = 4.23QDD1479 pKa = 3.56YY1480 pKa = 11.36SHH1482 pKa = 7.08GIVGKK1487 pKa = 8.32VLKK1490 pKa = 9.67LGPGIAPSSVTVTSDD1505 pKa = 2.7QYY1507 pKa = 11.58QDD1509 pKa = 3.34VVLTSGSDD1517 pKa = 3.46QIQLDD1522 pKa = 3.75GMADD1526 pKa = 3.35NSGLRR1531 pKa = 11.84GVQQVQFADD1540 pKa = 3.9GTLWTAAQMIAWARR1554 pKa = 11.84DD1555 pKa = 3.63VQGTPGNEE1563 pKa = 3.68TLYY1566 pKa = 9.5GTNGADD1572 pKa = 3.31IFDD1575 pKa = 4.24GQGGNDD1581 pKa = 3.18IEE1583 pKa = 4.7IGNGGADD1590 pKa = 2.76TFVFNQGYY1598 pKa = 8.21GQLEE1602 pKa = 3.89IDD1604 pKa = 3.8EE1605 pKa = 4.73ANYY1608 pKa = 10.65YY1609 pKa = 10.14PGTNSVLRR1617 pKa = 11.84LGAGIDD1623 pKa = 3.6EE1624 pKa = 5.03SEE1626 pKa = 4.46VSVAGTSSRR1635 pKa = 11.84GLVLTDD1641 pKa = 5.49GIAGDD1646 pKa = 5.45QITLDD1651 pKa = 3.44NALYY1655 pKa = 10.37GSDD1658 pKa = 3.61YY1659 pKa = 11.16GVQQVQFADD1668 pKa = 3.7GTTWSAQQLEE1678 pKa = 4.4QMATTGNAGGGTLYY1692 pKa = 8.34GTPGADD1698 pKa = 2.99VFDD1701 pKa = 4.16SRR1703 pKa = 11.84GGNATEE1709 pKa = 4.1VGNGGSDD1716 pKa = 3.4TYY1718 pKa = 11.34LLQPGYY1724 pKa = 11.22GPITIVNGISSNNVSEE1740 pKa = 4.47GVLSIAGVNPDD1751 pKa = 4.33NVWLAQVGNNLQVEE1765 pKa = 4.86VMGSTTEE1772 pKa = 3.8ATIQNWFNNSYY1783 pKa = 11.03SKK1785 pKa = 10.9LGEE1788 pKa = 4.06LTVSGGTAGNMTLDD1802 pKa = 3.42ARR1804 pKa = 11.84IDD1806 pKa = 3.67QLVQAMATFSSNNPGFDD1823 pKa = 3.67PTSSANPVINDD1834 pKa = 3.57PTLLTAVNSAWHH1846 pKa = 4.6QQ1847 pKa = 3.35
MM1 pKa = 8.08DD2 pKa = 4.31AQEE5 pKa = 4.8LGVLEE10 pKa = 4.87KK11 pKa = 10.28FTGEE15 pKa = 3.94NFVSSFTGGPDD26 pKa = 3.22PSNWHH31 pKa = 5.16QAQVLQDD38 pKa = 3.79AFNQLMNAVEE48 pKa = 4.27VRR50 pKa = 11.84LVAQGPLASYY60 pKa = 10.84LSGVGFNYY68 pKa = 9.13DD69 pKa = 3.04TDD71 pKa = 4.22SLVGSVDD78 pKa = 3.5LTALAEE84 pKa = 4.12ALINATPAGQLAAEE98 pKa = 5.45GYY100 pKa = 7.67WSSVAPMVGNLCSEE114 pKa = 4.82LGLGASAYY122 pKa = 7.54QTAFQGPFSSLNLPFSLDD140 pKa = 3.67EE141 pKa = 4.0IVQGNVFLGNGTNTVAVAHH160 pKa = 5.83TAGVPHH166 pKa = 6.65YY167 pKa = 9.9FDD169 pKa = 4.47GGPGVRR175 pKa = 11.84YY176 pKa = 9.36EE177 pKa = 4.27GSSGSGDD184 pKa = 3.02VFVFNQGYY192 pKa = 8.2GQLEE196 pKa = 4.05INEE199 pKa = 4.38FDD201 pKa = 3.37WNASNNVLRR210 pKa = 11.84LGAGITPDD218 pKa = 3.43AVKK221 pKa = 9.58VTLDD225 pKa = 3.73DD226 pKa = 4.0NQDD229 pKa = 3.01ILLTIGSNGDD239 pKa = 3.56QIKK242 pKa = 10.61LDD244 pKa = 3.85SEE246 pKa = 4.75GTGDD250 pKa = 4.02LRR252 pKa = 11.84WGVEE256 pKa = 3.89QVQFADD262 pKa = 4.13GAAWTQSQLIASARR276 pKa = 11.84NIQGTAASEE285 pKa = 4.05TLYY288 pKa = 9.62GTNGADD294 pKa = 3.2TFDD297 pKa = 4.52GKK299 pKa = 11.36GSTPYY304 pKa = 10.28SGSDD308 pKa = 3.36VEE310 pKa = 5.08IGNGGNDD317 pKa = 2.89TFIFNEE323 pKa = 5.21GYY325 pKa = 10.84GKK327 pKa = 8.74LTINEE332 pKa = 4.04YY333 pKa = 10.85DD334 pKa = 3.62YY335 pKa = 11.63GASPEE340 pKa = 4.15NVLQLGAGITPDD352 pKa = 3.32QVVVSVDD359 pKa = 3.04RR360 pKa = 11.84SQNIVLTIGTNGDD373 pKa = 3.05QVMLNRR379 pKa = 11.84EE380 pKa = 4.07ATGDD384 pKa = 3.39ARR386 pKa = 11.84EE387 pKa = 4.45GVQQIQFADD396 pKa = 3.94GTIWTQSQLLGMLATIDD413 pKa = 4.06GTPQDD418 pKa = 3.78DD419 pKa = 3.8TLYY422 pKa = 9.41GTGGANVFDD431 pKa = 4.41GKK433 pKa = 10.78GGYY436 pKa = 9.31DD437 pKa = 3.29VEE439 pKa = 4.63VGNGGNDD446 pKa = 3.13TYY448 pKa = 11.6VFNSGYY454 pKa = 8.15GTLVIKK460 pKa = 10.72DD461 pKa = 3.78SGSGVLDD468 pKa = 4.68LGTGITPSEE477 pKa = 4.19VKK479 pKa = 10.43ISTDD483 pKa = 3.25YY484 pKa = 11.55SGDD487 pKa = 3.21IVLTIGTEE495 pKa = 4.15GDD497 pKa = 3.42QVMLSGMTRR506 pKa = 11.84GASSSIQQVLFADD519 pKa = 4.59GTSWTAAQILAWSSDD534 pKa = 3.31IQGTTGNDD542 pKa = 3.33DD543 pKa = 4.12LYY545 pKa = 10.5GTSGDD550 pKa = 4.24DD551 pKa = 3.49VFDD554 pKa = 4.25GKK556 pKa = 11.08GGNDD560 pKa = 3.49YY561 pKa = 11.3EE562 pKa = 4.6SGRR565 pKa = 11.84GGNDD569 pKa = 2.83TFIFNQGYY577 pKa = 7.93GHH579 pKa = 7.37LEE581 pKa = 3.44ISEE584 pKa = 4.5VNGSAGFDD592 pKa = 3.41SVLEE596 pKa = 4.29LGSGITPSAVTIAYY610 pKa = 8.69GPNQSLVLTDD620 pKa = 4.41GNDD623 pKa = 3.24QITLDD628 pKa = 3.76GMANGSGRR636 pKa = 11.84DD637 pKa = 3.73GVQTVQFADD646 pKa = 3.9GTVWTASQMLASIGEE661 pKa = 4.06IRR663 pKa = 11.84GTAGDD668 pKa = 3.91DD669 pKa = 3.49QLFGTSGANLFDD681 pKa = 4.25GGGGNDD687 pKa = 3.82YY688 pKa = 11.36EE689 pKa = 6.34DD690 pKa = 4.83GDD692 pKa = 4.19GGNDD696 pKa = 3.06TFIFNRR702 pKa = 11.84GYY704 pKa = 8.6GTLEE708 pKa = 3.72IVEE711 pKa = 4.3QGAATTTGTLLLGPGILPSSVTVSLAGDD739 pKa = 3.88SVLLTIGSNGDD750 pKa = 3.66QVKK753 pKa = 10.4LDD755 pKa = 3.46YY756 pKa = 8.43MTDD759 pKa = 3.14RR760 pKa = 11.84GSNFGVRR767 pKa = 11.84DD768 pKa = 3.54VQFADD773 pKa = 3.73GTVWTASQILASLSDD788 pKa = 3.54VSGTAKK794 pKa = 10.36DD795 pKa = 3.8DD796 pKa = 4.62SIGGTSDD803 pKa = 3.27PSVIDD808 pKa = 3.75GKK810 pKa = 11.08GGNDD814 pKa = 3.14VAGGQGNNDD823 pKa = 3.27TFVFKK828 pKa = 10.72AGYY831 pKa = 9.4GHH833 pKa = 6.68LTVYY837 pKa = 10.11EE838 pKa = 4.03PQAANGVTNVLLFGPDD854 pKa = 2.87IDD856 pKa = 4.12EE857 pKa = 4.52SFVTVKK863 pKa = 10.91EE864 pKa = 4.01NMQDD868 pKa = 3.44AVVLTDD874 pKa = 5.28GIAGDD879 pKa = 4.13EE880 pKa = 4.23VTLVNQAYY888 pKa = 8.88EE889 pKa = 4.27FQGLQYY895 pKa = 11.13GGVQEE900 pKa = 4.31VQFADD905 pKa = 3.46GTMWTAQQLLQRR917 pKa = 11.84ATQGTPGDD925 pKa = 3.98DD926 pKa = 3.49VLIGGWQASLLDD938 pKa = 3.9GKK940 pKa = 10.9GGNDD944 pKa = 3.49YY945 pKa = 10.96EE946 pKa = 5.53AGGGDD951 pKa = 3.35GDD953 pKa = 3.83TFVFNAGYY961 pKa = 10.35GHH963 pKa = 6.7LQINEE968 pKa = 4.32YY969 pKa = 9.85QWGTANNVLQLGQGISEE986 pKa = 4.31SSISASVTRR995 pKa = 11.84GDD997 pKa = 5.25AIVLSDD1003 pKa = 4.41GTPGDD1008 pKa = 3.79QIVLDD1013 pKa = 4.44GEE1015 pKa = 4.33FDD1017 pKa = 3.56DD1018 pKa = 5.2ARR1020 pKa = 11.84YY1021 pKa = 10.21GGTSRR1026 pKa = 11.84GIQEE1030 pKa = 3.89VQFADD1035 pKa = 3.51GTTWSVQQLVQLAQTRR1051 pKa = 11.84TGTPGDD1057 pKa = 4.23DD1058 pKa = 3.62YY1059 pKa = 11.91LVGTTTASVFDD1070 pKa = 4.15GKK1072 pKa = 10.93GGNDD1076 pKa = 3.35FEE1078 pKa = 4.87TGGGKK1083 pKa = 10.5GDD1085 pKa = 3.32TFVFNAGYY1093 pKa = 9.82GHH1095 pKa = 7.08LEE1097 pKa = 3.9IYY1099 pKa = 9.18EE1100 pKa = 4.1YY1101 pKa = 9.92PYY1103 pKa = 10.97QPTNVLQLGPGISASSVEE1121 pKa = 4.29VSATEE1126 pKa = 4.08DD1127 pKa = 3.4YY1128 pKa = 11.41GVILTDD1134 pKa = 3.78GMAGDD1139 pKa = 4.71EE1140 pKa = 4.29IEE1142 pKa = 5.36LDD1144 pKa = 3.8GEE1146 pKa = 4.39LGGEE1150 pKa = 4.26GVEE1153 pKa = 4.01QVQFSDD1159 pKa = 3.48GTVWTRR1165 pKa = 11.84QQLLQMAEE1173 pKa = 4.39TTTGTPGDD1181 pKa = 4.16DD1182 pKa = 3.53VLLGTTDD1189 pKa = 3.41ASLFDD1194 pKa = 3.95GKK1196 pKa = 10.95GGNDD1200 pKa = 3.46YY1201 pKa = 11.6EE1202 pKa = 4.39EE1203 pKa = 5.86GDD1205 pKa = 4.26GNGHH1209 pKa = 7.5LDD1211 pKa = 3.29TFVFNAGYY1219 pKa = 9.93GHH1221 pKa = 7.63LEE1223 pKa = 3.8IDD1225 pKa = 3.46EE1226 pKa = 4.55SNYY1229 pKa = 9.7TVATSVLEE1237 pKa = 4.05LGPGISEE1244 pKa = 4.46SSLQVTTDD1252 pKa = 2.93KK1253 pKa = 11.64SEE1255 pKa = 5.36DD1256 pKa = 3.34IVLTSGTDD1264 pKa = 3.2QIVIDD1269 pKa = 4.21WMADD1273 pKa = 2.67GWGDD1277 pKa = 3.63YY1278 pKa = 11.07GVGQVTFADD1287 pKa = 4.17GTVWTASQIIAMASNVQGTAGNDD1310 pKa = 3.44TLYY1313 pKa = 9.57GTSISSTFDD1322 pKa = 3.02GKK1324 pKa = 11.04GGNDD1328 pKa = 3.29VEE1330 pKa = 4.66IGNGNDD1336 pKa = 3.12DD1337 pKa = 3.87TFIFDD1342 pKa = 3.78PGYY1345 pKa = 9.3GHH1347 pKa = 7.66LEE1349 pKa = 3.98IEE1351 pKa = 4.4EE1352 pKa = 4.28GMANYY1357 pKa = 10.64DD1358 pKa = 3.63STNVLQFGAGIRR1370 pKa = 11.84AAMVTASLGSNGSIVLTSGTDD1391 pKa = 3.43QVVLDD1396 pKa = 3.97GMTTQSCGIQLVQFADD1412 pKa = 4.14GTQWTPSQILAMASIIRR1429 pKa = 11.84GTAGNDD1435 pKa = 3.63TLSGTAGGDD1444 pKa = 3.05WFDD1447 pKa = 3.71GRR1449 pKa = 11.84GGNDD1453 pKa = 3.39LEE1455 pKa = 4.58VGGGGADD1462 pKa = 2.76TFVFNQGYY1470 pKa = 8.16GHH1472 pKa = 7.39LEE1474 pKa = 3.64IDD1476 pKa = 3.83EE1477 pKa = 4.23QDD1479 pKa = 3.56YY1480 pKa = 11.36SHH1482 pKa = 7.08GIVGKK1487 pKa = 8.32VLKK1490 pKa = 9.67LGPGIAPSSVTVTSDD1505 pKa = 2.7QYY1507 pKa = 11.58QDD1509 pKa = 3.34VVLTSGSDD1517 pKa = 3.46QIQLDD1522 pKa = 3.75GMADD1526 pKa = 3.35NSGLRR1531 pKa = 11.84GVQQVQFADD1540 pKa = 3.9GTLWTAAQMIAWARR1554 pKa = 11.84DD1555 pKa = 3.63VQGTPGNEE1563 pKa = 3.68TLYY1566 pKa = 9.5GTNGADD1572 pKa = 3.31IFDD1575 pKa = 4.24GQGGNDD1581 pKa = 3.18IEE1583 pKa = 4.7IGNGGADD1590 pKa = 2.76TFVFNQGYY1598 pKa = 8.21GQLEE1602 pKa = 3.89IDD1604 pKa = 3.8EE1605 pKa = 4.73ANYY1608 pKa = 10.65YY1609 pKa = 10.14PGTNSVLRR1617 pKa = 11.84LGAGIDD1623 pKa = 3.6EE1624 pKa = 5.03SEE1626 pKa = 4.46VSVAGTSSRR1635 pKa = 11.84GLVLTDD1641 pKa = 5.49GIAGDD1646 pKa = 5.45QITLDD1651 pKa = 3.44NALYY1655 pKa = 10.37GSDD1658 pKa = 3.61YY1659 pKa = 11.16GVQQVQFADD1668 pKa = 3.7GTTWSAQQLEE1678 pKa = 4.4QMATTGNAGGGTLYY1692 pKa = 8.34GTPGADD1698 pKa = 2.99VFDD1701 pKa = 4.16SRR1703 pKa = 11.84GGNATEE1709 pKa = 4.1VGNGGSDD1716 pKa = 3.4TYY1718 pKa = 11.34LLQPGYY1724 pKa = 11.22GPITIVNGISSNNVSEE1740 pKa = 4.47GVLSIAGVNPDD1751 pKa = 4.33NVWLAQVGNNLQVEE1765 pKa = 4.86VMGSTTEE1772 pKa = 3.8ATIQNWFNNSYY1783 pKa = 11.03SKK1785 pKa = 10.9LGEE1788 pKa = 4.06LTVSGGTAGNMTLDD1802 pKa = 3.42ARR1804 pKa = 11.84IDD1806 pKa = 3.67QLVQAMATFSSNNPGFDD1823 pKa = 3.67PTSSANPVINDD1834 pKa = 3.57PTLLTAVNSAWHH1846 pKa = 4.6QQ1847 pKa = 3.35
Molecular weight: 191.62 kDa
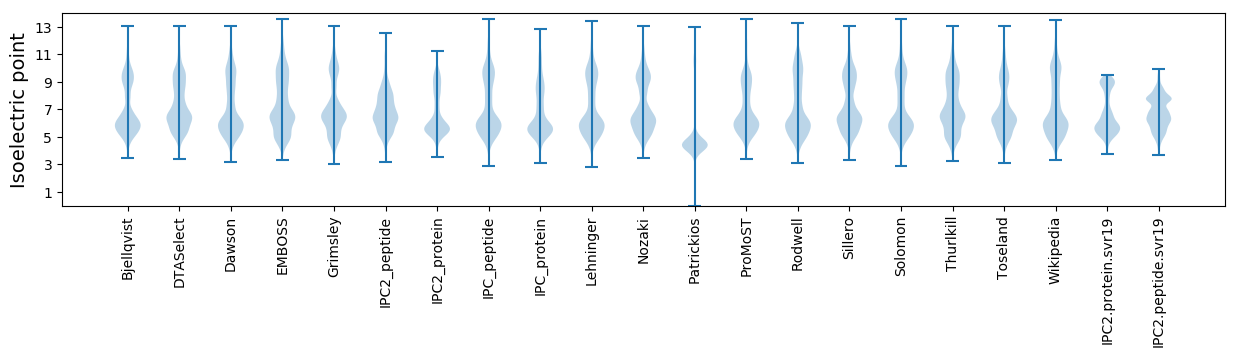
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N7X5M2|A0A2N7X5M2_9BURK Nuclear transport factor 2 family protein OS=Trinickia symbiotica OX=863227 GN=C0Z20_11780 PE=4 SV=1
MM1 pKa = 6.8TASARR6 pKa = 11.84TAGTASAGTTSGTASGTTGGTSGTASGTTGTTGTTGTTGTTSGTTSGTTSGTTSARR62 pKa = 11.84TAGARR67 pKa = 11.84TAGARR72 pKa = 11.84TAGARR77 pKa = 11.84TASARR82 pKa = 11.84TAGARR87 pKa = 11.84TASAGTASAGTTSGTTSARR106 pKa = 11.84TASTGTASTRR116 pKa = 11.84TASTRR121 pKa = 11.84TASTRR126 pKa = 11.84TASAASTRR134 pKa = 11.84TAAA137 pKa = 4.39
MM1 pKa = 6.8TASARR6 pKa = 11.84TAGTASAGTTSGTASGTTGGTSGTASGTTGTTGTTGTTGTTSGTTSGTTSGTTSARR62 pKa = 11.84TAGARR67 pKa = 11.84TAGARR72 pKa = 11.84TAGARR77 pKa = 11.84TASARR82 pKa = 11.84TAGARR87 pKa = 11.84TASAGTASAGTTSGTTSARR106 pKa = 11.84TASTGTASTRR116 pKa = 11.84TASTRR121 pKa = 11.84TASTRR126 pKa = 11.84TASAASTRR134 pKa = 11.84TAAA137 pKa = 4.39
Molecular weight: 12.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1928138 |
23 |
3089 |
322.3 |
34.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.874 ± 0.046 | 0.96 ± 0.011 |
5.395 ± 0.023 | 5.436 ± 0.036 |
3.671 ± 0.019 | 8.208 ± 0.035 |
2.277 ± 0.016 | 4.838 ± 0.023 |
3.014 ± 0.029 | 10.056 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.312 ± 0.015 | 2.712 ± 0.022 |
5.092 ± 0.024 | 3.32 ± 0.023 |
7.246 ± 0.038 | 5.731 ± 0.028 |
5.336 ± 0.03 | 7.728 ± 0.024 |
1.356 ± 0.013 | 2.438 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
