
Bacillus daliensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalicoccus
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
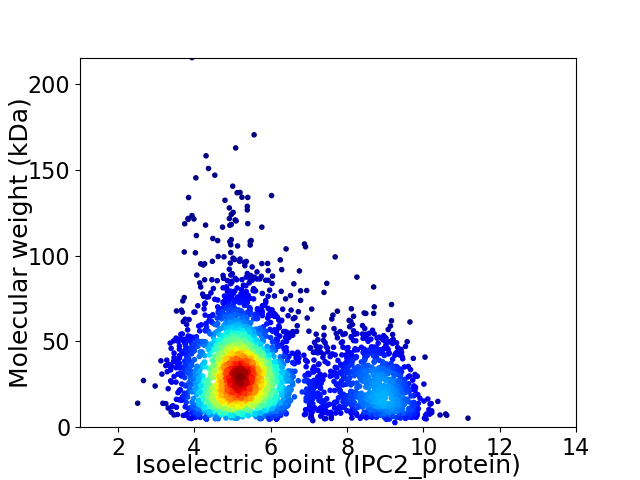
Virtual 2D-PAGE plot for 3459 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0GAP2|A0A1H0GAP2_9BACI Multiple sugar transport system permease protein OS=Bacillus daliensis OX=745820 GN=SAMN04488053_10637 PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 10.1KK3 pKa = 10.31KK4 pKa = 10.52KK5 pKa = 9.77FLSSILGVALVGVIAGCGGSDD26 pKa = 4.55NNDD29 pKa = 2.88NTLNDD34 pKa = 3.64TAEE37 pKa = 4.18GSEE40 pKa = 4.22VNNAGAEE47 pKa = 4.12AEE49 pKa = 4.27EE50 pKa = 4.12EE51 pKa = 4.1AVTIRR56 pKa = 11.84LHH58 pKa = 4.77HH59 pKa = 7.1WYY61 pKa = 10.48NIEE64 pKa = 3.6QDD66 pKa = 2.97NWNEE70 pKa = 3.47VVAAFEE76 pKa = 4.42EE77 pKa = 4.51EE78 pKa = 4.29HH79 pKa = 5.77PHH81 pKa = 7.78INIEE85 pKa = 4.21MNSPEE90 pKa = 4.23NNDD93 pKa = 3.27ANEE96 pKa = 4.37TVQQMDD102 pKa = 4.14LAAASGDD109 pKa = 3.62QLDD112 pKa = 3.86VMMINNAQDD121 pKa = 3.13YY122 pKa = 7.81SQRR125 pKa = 11.84VGQGMFEE132 pKa = 4.21PLNGFIEE139 pKa = 4.42EE140 pKa = 4.61DD141 pKa = 3.71GYY143 pKa = 11.46EE144 pKa = 4.13YY145 pKa = 10.8EE146 pKa = 5.75DD147 pKa = 4.16EE148 pKa = 4.61YY149 pKa = 11.8SVNTAVDD156 pKa = 3.37GDD158 pKa = 4.58YY159 pKa = 11.2YY160 pKa = 11.55ALPGKK165 pKa = 10.47FNGFFVMLNEE175 pKa = 4.98DD176 pKa = 3.98ALDD179 pKa = 3.9EE180 pKa = 5.04AGLEE184 pKa = 4.43VPEE187 pKa = 4.01EE188 pKa = 4.38WTWDD192 pKa = 3.22EE193 pKa = 4.09YY194 pKa = 11.01MDD196 pKa = 3.9YY197 pKa = 11.31AEE199 pKa = 4.55QLSEE203 pKa = 4.69GEE205 pKa = 4.37GADD208 pKa = 3.28RR209 pKa = 11.84RR210 pKa = 11.84YY211 pKa = 9.7GTFFHH216 pKa = 6.47TWLDD220 pKa = 3.71YY221 pKa = 10.77MKK223 pKa = 10.56LAVFNQPEE231 pKa = 4.17DD232 pKa = 3.89SNIIKK237 pKa = 10.44DD238 pKa = 4.16DD239 pKa = 3.84GVTSNIDD246 pKa = 3.25SEE248 pKa = 4.86HH249 pKa = 5.78IRR251 pKa = 11.84KK252 pKa = 9.51SLEE255 pKa = 3.22IRR257 pKa = 11.84EE258 pKa = 4.46RR259 pKa = 11.84GHH261 pKa = 6.8EE262 pKa = 4.38DD263 pKa = 3.3NSATPYY269 pKa = 11.22ADD271 pKa = 4.14TISLDD276 pKa = 3.51LNYY279 pKa = 9.93RR280 pKa = 11.84DD281 pKa = 3.59QFFGEE286 pKa = 4.32SAAMIMTGTWMINDD300 pKa = 3.56TSGTDD305 pKa = 3.33EE306 pKa = 4.27YY307 pKa = 10.85PPFNLAYY314 pKa = 10.23APYY317 pKa = 8.93PKK319 pKa = 10.19AAEE322 pKa = 4.73GDD324 pKa = 4.08DD325 pKa = 3.78STSPSGADD333 pKa = 3.21FLAIYY338 pKa = 10.15SGSDD342 pKa = 3.45YY343 pKa = 11.14KK344 pKa = 11.25EE345 pKa = 3.52EE346 pKa = 4.47AYY348 pKa = 10.25EE349 pKa = 4.18FIRR352 pKa = 11.84WYY354 pKa = 6.54TTEE357 pKa = 4.82GITMQGKK364 pKa = 9.03YY365 pKa = 10.5LPDD368 pKa = 3.88PQAVDD373 pKa = 2.92IEE375 pKa = 4.47EE376 pKa = 4.33VVNNLLEE383 pKa = 4.34EE384 pKa = 5.03ADD386 pKa = 5.16DD387 pKa = 4.55LDD389 pKa = 4.28QEE391 pKa = 4.67GVNVDD396 pKa = 3.87SLIHH400 pKa = 5.23TLEE403 pKa = 4.18TQEE406 pKa = 4.16PAALNIPPAYY416 pKa = 9.43IGEE419 pKa = 4.3AEE421 pKa = 4.02SAYY424 pKa = 9.79EE425 pKa = 3.93NEE427 pKa = 4.38VEE429 pKa = 5.06SFLLGDD435 pKa = 3.66QDD437 pKa = 5.58LDD439 pKa = 3.86TTIEE443 pKa = 4.07NAHH446 pKa = 6.3EE447 pKa = 4.07AVQNVIDD454 pKa = 4.06QNAEE458 pKa = 3.59
MM1 pKa = 7.54KK2 pKa = 10.1KK3 pKa = 10.31KK4 pKa = 10.52KK5 pKa = 9.77FLSSILGVALVGVIAGCGGSDD26 pKa = 4.55NNDD29 pKa = 2.88NTLNDD34 pKa = 3.64TAEE37 pKa = 4.18GSEE40 pKa = 4.22VNNAGAEE47 pKa = 4.12AEE49 pKa = 4.27EE50 pKa = 4.12EE51 pKa = 4.1AVTIRR56 pKa = 11.84LHH58 pKa = 4.77HH59 pKa = 7.1WYY61 pKa = 10.48NIEE64 pKa = 3.6QDD66 pKa = 2.97NWNEE70 pKa = 3.47VVAAFEE76 pKa = 4.42EE77 pKa = 4.51EE78 pKa = 4.29HH79 pKa = 5.77PHH81 pKa = 7.78INIEE85 pKa = 4.21MNSPEE90 pKa = 4.23NNDD93 pKa = 3.27ANEE96 pKa = 4.37TVQQMDD102 pKa = 4.14LAAASGDD109 pKa = 3.62QLDD112 pKa = 3.86VMMINNAQDD121 pKa = 3.13YY122 pKa = 7.81SQRR125 pKa = 11.84VGQGMFEE132 pKa = 4.21PLNGFIEE139 pKa = 4.42EE140 pKa = 4.61DD141 pKa = 3.71GYY143 pKa = 11.46EE144 pKa = 4.13YY145 pKa = 10.8EE146 pKa = 5.75DD147 pKa = 4.16EE148 pKa = 4.61YY149 pKa = 11.8SVNTAVDD156 pKa = 3.37GDD158 pKa = 4.58YY159 pKa = 11.2YY160 pKa = 11.55ALPGKK165 pKa = 10.47FNGFFVMLNEE175 pKa = 4.98DD176 pKa = 3.98ALDD179 pKa = 3.9EE180 pKa = 5.04AGLEE184 pKa = 4.43VPEE187 pKa = 4.01EE188 pKa = 4.38WTWDD192 pKa = 3.22EE193 pKa = 4.09YY194 pKa = 11.01MDD196 pKa = 3.9YY197 pKa = 11.31AEE199 pKa = 4.55QLSEE203 pKa = 4.69GEE205 pKa = 4.37GADD208 pKa = 3.28RR209 pKa = 11.84RR210 pKa = 11.84YY211 pKa = 9.7GTFFHH216 pKa = 6.47TWLDD220 pKa = 3.71YY221 pKa = 10.77MKK223 pKa = 10.56LAVFNQPEE231 pKa = 4.17DD232 pKa = 3.89SNIIKK237 pKa = 10.44DD238 pKa = 4.16DD239 pKa = 3.84GVTSNIDD246 pKa = 3.25SEE248 pKa = 4.86HH249 pKa = 5.78IRR251 pKa = 11.84KK252 pKa = 9.51SLEE255 pKa = 3.22IRR257 pKa = 11.84EE258 pKa = 4.46RR259 pKa = 11.84GHH261 pKa = 6.8EE262 pKa = 4.38DD263 pKa = 3.3NSATPYY269 pKa = 11.22ADD271 pKa = 4.14TISLDD276 pKa = 3.51LNYY279 pKa = 9.93RR280 pKa = 11.84DD281 pKa = 3.59QFFGEE286 pKa = 4.32SAAMIMTGTWMINDD300 pKa = 3.56TSGTDD305 pKa = 3.33EE306 pKa = 4.27YY307 pKa = 10.85PPFNLAYY314 pKa = 10.23APYY317 pKa = 8.93PKK319 pKa = 10.19AAEE322 pKa = 4.73GDD324 pKa = 4.08DD325 pKa = 3.78STSPSGADD333 pKa = 3.21FLAIYY338 pKa = 10.15SGSDD342 pKa = 3.45YY343 pKa = 11.14KK344 pKa = 11.25EE345 pKa = 3.52EE346 pKa = 4.47AYY348 pKa = 10.25EE349 pKa = 4.18FIRR352 pKa = 11.84WYY354 pKa = 6.54TTEE357 pKa = 4.82GITMQGKK364 pKa = 9.03YY365 pKa = 10.5LPDD368 pKa = 3.88PQAVDD373 pKa = 2.92IEE375 pKa = 4.47EE376 pKa = 4.33VVNNLLEE383 pKa = 4.34EE384 pKa = 5.03ADD386 pKa = 5.16DD387 pKa = 4.55LDD389 pKa = 4.28QEE391 pKa = 4.67GVNVDD396 pKa = 3.87SLIHH400 pKa = 5.23TLEE403 pKa = 4.18TQEE406 pKa = 4.16PAALNIPPAYY416 pKa = 9.43IGEE419 pKa = 4.3AEE421 pKa = 4.02SAYY424 pKa = 9.79EE425 pKa = 3.93NEE427 pKa = 4.38VEE429 pKa = 5.06SFLLGDD435 pKa = 3.66QDD437 pKa = 5.58LDD439 pKa = 3.86TTIEE443 pKa = 4.07NAHH446 pKa = 6.3EE447 pKa = 4.07AVQNVIDD454 pKa = 4.06QNAEE458 pKa = 3.59
Molecular weight: 51.21 kDa
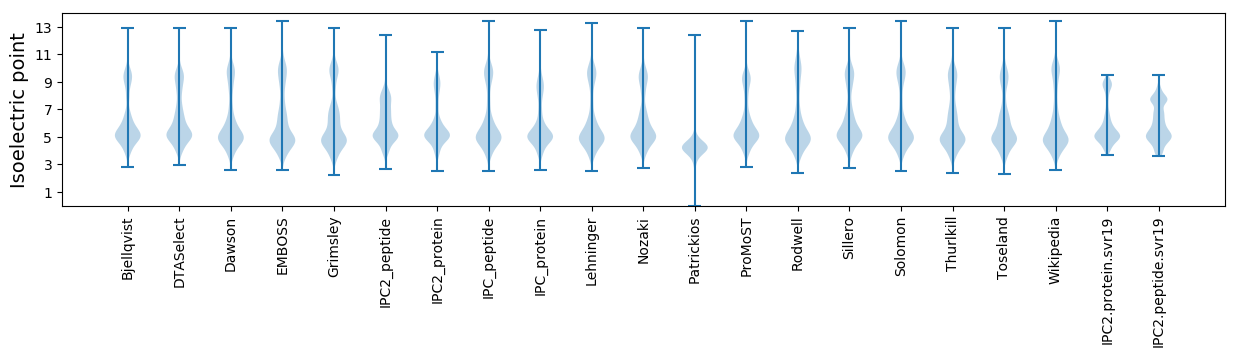
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0FN76|A0A1H0FN76_9BACI DNA replication and repair protein RecF OS=Bacillus daliensis OX=745820 GN=recF PE=3 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.34KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.17VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.34RR13 pKa = 11.84KK14 pKa = 8.34KK15 pKa = 9.1NHH17 pKa = 4.73GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37NGRR29 pKa = 11.84KK30 pKa = 9.17VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1040143 |
25 |
1926 |
300.7 |
33.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.932 ± 0.037 | 0.603 ± 0.011 |
5.137 ± 0.036 | 8.805 ± 0.065 |
4.432 ± 0.035 | 6.955 ± 0.042 |
2.161 ± 0.021 | 7.084 ± 0.04 |
6.067 ± 0.047 | 9.542 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.899 ± 0.018 | 4.083 ± 0.037 |
3.701 ± 0.023 | 3.796 ± 0.027 |
4.195 ± 0.026 | 5.945 ± 0.034 |
5.38 ± 0.026 | 6.851 ± 0.037 |
1.067 ± 0.015 | 3.365 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
