
Nocardiopsis alba (strain ATCC BAA-2165 / BE74)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Nocardiopsis; Nocardiopsis alba
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
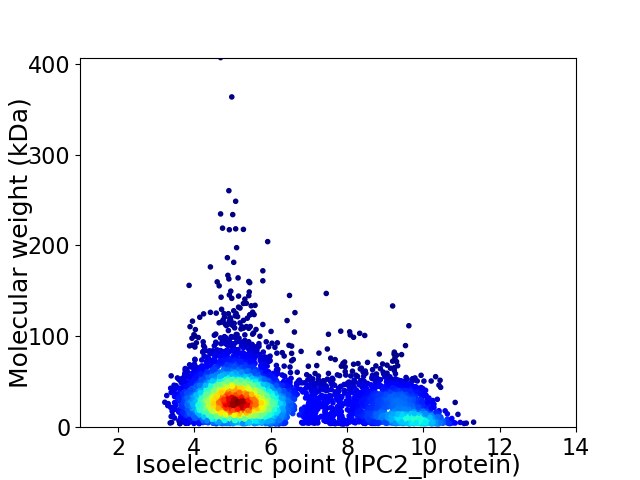
Virtual 2D-PAGE plot for 5500 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7LIH7|J7LIH7_NOCAA Branched-chain amino acid transport system / permease component family protein OS=Nocardiopsis alba (strain ATCC BAA-2165 / BE74) OX=1205910 GN=B005_3878 PE=4 SV=1
MM1 pKa = 7.49IKK3 pKa = 10.35GVTMFEE9 pKa = 3.98RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 8.19TLALAAAATSTVLVVGGCGSGDD34 pKa = 3.33GGGGDD39 pKa = 3.86GGPKK43 pKa = 9.48EE44 pKa = 4.37VTWVINSLPGAWQATSLAGGSVYY67 pKa = 10.15VVQMLSGVLPYY78 pKa = 9.97TGQWQPDD85 pKa = 3.36GTYY88 pKa = 9.86EE89 pKa = 4.23YY90 pKa = 11.25DD91 pKa = 3.48MNVLAEE97 pKa = 4.25EE98 pKa = 4.78PEE100 pKa = 5.01LINDD104 pKa = 4.82DD105 pKa = 3.88PDD107 pKa = 3.56EE108 pKa = 5.39GPFEE112 pKa = 4.09WSITLAEE119 pKa = 4.28DD120 pKa = 4.06AVWSDD125 pKa = 3.73GEE127 pKa = 4.3PMTGEE132 pKa = 4.31DD133 pKa = 5.37LRR135 pKa = 11.84VSWMLGTSPDD145 pKa = 3.11EE146 pKa = 4.98GYY148 pKa = 10.91CGGCDD153 pKa = 3.3PRR155 pKa = 11.84AAASFDD161 pKa = 3.37KK162 pKa = 11.23VEE164 pKa = 4.25SVEE167 pKa = 4.54ADD169 pKa = 2.92GKK171 pKa = 8.97TATFQLKK178 pKa = 10.2EE179 pKa = 4.0GLADD183 pKa = 4.08PEE185 pKa = 4.31WMSLFDD191 pKa = 3.83AHH193 pKa = 6.22SQSGGFLPAHH203 pKa = 6.49LAEE206 pKa = 4.4EE207 pKa = 4.52NGWDD211 pKa = 4.83VDD213 pKa = 4.61DD214 pKa = 5.61ADD216 pKa = 4.86QLGEE220 pKa = 4.18YY221 pKa = 10.01YY222 pKa = 10.64DD223 pKa = 3.57WLHH226 pKa = 6.76AEE228 pKa = 4.08RR229 pKa = 11.84PEE231 pKa = 4.25WSGGPYY237 pKa = 9.74MITAGDD243 pKa = 3.95LEE245 pKa = 4.58NEE247 pKa = 4.61VVKK250 pKa = 10.64EE251 pKa = 3.86PNPNYY256 pKa = 10.33FGEE259 pKa = 4.45EE260 pKa = 3.88PLLDD264 pKa = 5.26KK265 pKa = 10.39ITMPYY270 pKa = 8.42NTDD273 pKa = 2.65EE274 pKa = 4.34GTFVNAFINGEE285 pKa = 3.85IDD287 pKa = 3.17GGNPADD293 pKa = 3.87YY294 pKa = 10.93SEE296 pKa = 5.46DD297 pKa = 3.94VILQLQDD304 pKa = 3.13VANAEE309 pKa = 4.06VTIAEE314 pKa = 4.49GATWEE319 pKa = 4.59HH320 pKa = 6.46VDD322 pKa = 5.03LNLKK326 pKa = 10.38NEE328 pKa = 4.16TLQDD332 pKa = 3.44VEE334 pKa = 4.07LRR336 pKa = 11.84RR337 pKa = 11.84AIFTAIDD344 pKa = 3.45RR345 pKa = 11.84DD346 pKa = 4.37DD347 pKa = 3.41IANRR351 pKa = 11.84NFGAGYY357 pKa = 9.49PDD359 pKa = 4.23YY360 pKa = 10.47EE361 pKa = 4.63LKK363 pKa = 10.82NNHH366 pKa = 5.4VFGSDD371 pKa = 2.76SPYY374 pKa = 10.92YY375 pKa = 9.3VDD377 pKa = 3.96HH378 pKa = 7.46FEE380 pKa = 5.86GEE382 pKa = 4.42TQGTGDD388 pKa = 3.32IDD390 pKa = 3.67EE391 pKa = 4.99AKK393 pKa = 10.66SILEE397 pKa = 3.91EE398 pKa = 4.6AGYY401 pKa = 10.42EE402 pKa = 3.97LDD404 pKa = 5.27GGTLTLDD411 pKa = 3.78GEE413 pKa = 4.81EE414 pKa = 4.03IGPFRR419 pKa = 11.84LRR421 pKa = 11.84STDD424 pKa = 2.99STVRR428 pKa = 11.84NTSVQLIQAQLSEE441 pKa = 4.28IGITTNIEE449 pKa = 3.83MTDD452 pKa = 3.57DD453 pKa = 3.87LGGMLAAAEE462 pKa = 4.09YY463 pKa = 10.74DD464 pKa = 3.09IVQYY468 pKa = 10.53GWSGSPYY475 pKa = 8.93FTGNPDD481 pKa = 3.61QFWHH485 pKa = 6.38SEE487 pKa = 3.93SGSNFGGYY495 pKa = 10.48SNDD498 pKa = 3.74EE499 pKa = 4.05VDD501 pKa = 5.55DD502 pKa = 4.97LADD505 pKa = 3.53QVANAPDD512 pKa = 4.51LDD514 pKa = 4.17TAAEE518 pKa = 4.07HH519 pKa = 6.81ANAAMEE525 pKa = 4.09ILVPEE530 pKa = 4.94AYY532 pKa = 9.98VLPIMAEE539 pKa = 3.82PNYY542 pKa = 10.2FFANSDD548 pKa = 3.55RR549 pKa = 11.84LVNVHH554 pKa = 7.4DD555 pKa = 4.7NLQSSYY561 pKa = 10.71RR562 pKa = 11.84ATYY565 pKa = 10.4NIGEE569 pKa = 4.22WDD571 pKa = 3.58VAGG574 pKa = 4.12
MM1 pKa = 7.49IKK3 pKa = 10.35GVTMFEE9 pKa = 3.98RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 8.19TLALAAAATSTVLVVGGCGSGDD34 pKa = 3.33GGGGDD39 pKa = 3.86GGPKK43 pKa = 9.48EE44 pKa = 4.37VTWVINSLPGAWQATSLAGGSVYY67 pKa = 10.15VVQMLSGVLPYY78 pKa = 9.97TGQWQPDD85 pKa = 3.36GTYY88 pKa = 9.86EE89 pKa = 4.23YY90 pKa = 11.25DD91 pKa = 3.48MNVLAEE97 pKa = 4.25EE98 pKa = 4.78PEE100 pKa = 5.01LINDD104 pKa = 4.82DD105 pKa = 3.88PDD107 pKa = 3.56EE108 pKa = 5.39GPFEE112 pKa = 4.09WSITLAEE119 pKa = 4.28DD120 pKa = 4.06AVWSDD125 pKa = 3.73GEE127 pKa = 4.3PMTGEE132 pKa = 4.31DD133 pKa = 5.37LRR135 pKa = 11.84VSWMLGTSPDD145 pKa = 3.11EE146 pKa = 4.98GYY148 pKa = 10.91CGGCDD153 pKa = 3.3PRR155 pKa = 11.84AAASFDD161 pKa = 3.37KK162 pKa = 11.23VEE164 pKa = 4.25SVEE167 pKa = 4.54ADD169 pKa = 2.92GKK171 pKa = 8.97TATFQLKK178 pKa = 10.2EE179 pKa = 4.0GLADD183 pKa = 4.08PEE185 pKa = 4.31WMSLFDD191 pKa = 3.83AHH193 pKa = 6.22SQSGGFLPAHH203 pKa = 6.49LAEE206 pKa = 4.4EE207 pKa = 4.52NGWDD211 pKa = 4.83VDD213 pKa = 4.61DD214 pKa = 5.61ADD216 pKa = 4.86QLGEE220 pKa = 4.18YY221 pKa = 10.01YY222 pKa = 10.64DD223 pKa = 3.57WLHH226 pKa = 6.76AEE228 pKa = 4.08RR229 pKa = 11.84PEE231 pKa = 4.25WSGGPYY237 pKa = 9.74MITAGDD243 pKa = 3.95LEE245 pKa = 4.58NEE247 pKa = 4.61VVKK250 pKa = 10.64EE251 pKa = 3.86PNPNYY256 pKa = 10.33FGEE259 pKa = 4.45EE260 pKa = 3.88PLLDD264 pKa = 5.26KK265 pKa = 10.39ITMPYY270 pKa = 8.42NTDD273 pKa = 2.65EE274 pKa = 4.34GTFVNAFINGEE285 pKa = 3.85IDD287 pKa = 3.17GGNPADD293 pKa = 3.87YY294 pKa = 10.93SEE296 pKa = 5.46DD297 pKa = 3.94VILQLQDD304 pKa = 3.13VANAEE309 pKa = 4.06VTIAEE314 pKa = 4.49GATWEE319 pKa = 4.59HH320 pKa = 6.46VDD322 pKa = 5.03LNLKK326 pKa = 10.38NEE328 pKa = 4.16TLQDD332 pKa = 3.44VEE334 pKa = 4.07LRR336 pKa = 11.84RR337 pKa = 11.84AIFTAIDD344 pKa = 3.45RR345 pKa = 11.84DD346 pKa = 4.37DD347 pKa = 3.41IANRR351 pKa = 11.84NFGAGYY357 pKa = 9.49PDD359 pKa = 4.23YY360 pKa = 10.47EE361 pKa = 4.63LKK363 pKa = 10.82NNHH366 pKa = 5.4VFGSDD371 pKa = 2.76SPYY374 pKa = 10.92YY375 pKa = 9.3VDD377 pKa = 3.96HH378 pKa = 7.46FEE380 pKa = 5.86GEE382 pKa = 4.42TQGTGDD388 pKa = 3.32IDD390 pKa = 3.67EE391 pKa = 4.99AKK393 pKa = 10.66SILEE397 pKa = 3.91EE398 pKa = 4.6AGYY401 pKa = 10.42EE402 pKa = 3.97LDD404 pKa = 5.27GGTLTLDD411 pKa = 3.78GEE413 pKa = 4.81EE414 pKa = 4.03IGPFRR419 pKa = 11.84LRR421 pKa = 11.84STDD424 pKa = 2.99STVRR428 pKa = 11.84NTSVQLIQAQLSEE441 pKa = 4.28IGITTNIEE449 pKa = 3.83MTDD452 pKa = 3.57DD453 pKa = 3.87LGGMLAAAEE462 pKa = 4.09YY463 pKa = 10.74DD464 pKa = 3.09IVQYY468 pKa = 10.53GWSGSPYY475 pKa = 8.93FTGNPDD481 pKa = 3.61QFWHH485 pKa = 6.38SEE487 pKa = 3.93SGSNFGGYY495 pKa = 10.48SNDD498 pKa = 3.74EE499 pKa = 4.05VDD501 pKa = 5.55DD502 pKa = 4.97LADD505 pKa = 3.53QVANAPDD512 pKa = 4.51LDD514 pKa = 4.17TAAEE518 pKa = 4.07HH519 pKa = 6.81ANAAMEE525 pKa = 4.09ILVPEE530 pKa = 4.94AYY532 pKa = 9.98VLPIMAEE539 pKa = 3.82PNYY542 pKa = 10.2FFANSDD548 pKa = 3.55RR549 pKa = 11.84LVNVHH554 pKa = 7.4DD555 pKa = 4.7NLQSSYY561 pKa = 10.71RR562 pKa = 11.84ATYY565 pKa = 10.4NIGEE569 pKa = 4.22WDD571 pKa = 3.58VAGG574 pKa = 4.12
Molecular weight: 62.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7L8U2|J7L8U2_NOCAA YcfA-like family protein OS=Nocardiopsis alba (strain ATCC BAA-2165 / BE74) OX=1205910 GN=B005_2467 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.5GRR40 pKa = 11.84AALTVSHH47 pKa = 7.07
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.45VHH17 pKa = 5.31GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.5GRR40 pKa = 11.84AALTVSHH47 pKa = 7.07
Molecular weight: 5.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1675371 |
37 |
3832 |
304.6 |
32.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.691 ± 0.042 | 0.707 ± 0.009 |
6.261 ± 0.034 | 7.046 ± 0.037 |
2.834 ± 0.02 | 9.527 ± 0.037 |
2.336 ± 0.017 | 3.485 ± 0.026 |
1.617 ± 0.029 | 10.383 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.971 ± 0.014 | 1.744 ± 0.019 |
6.12 ± 0.035 | 2.149 ± 0.018 |
8.587 ± 0.038 | 5.521 ± 0.023 |
5.915 ± 0.027 | 8.679 ± 0.034 |
1.472 ± 0.015 | 1.956 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
