
Helicoverpa zea nudivirus 2 (HzNV-2)
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Nudiviridae; Betanudivirus; Heliothis zea nudivirus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
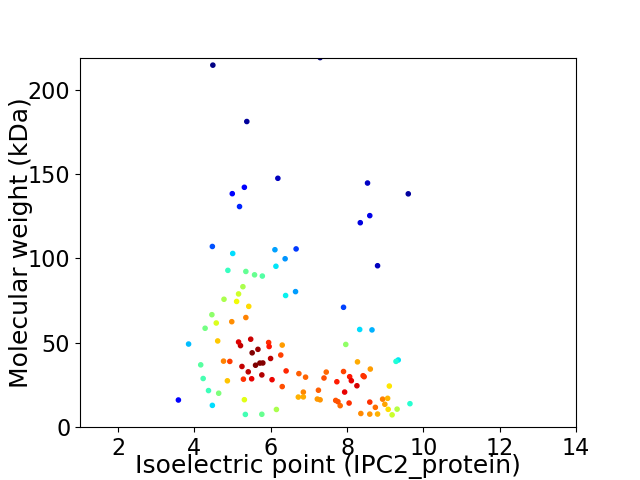
Virtual 2D-PAGE plot for 113 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9I040|G9I040_HZNV2 Uncharacterized protein orf14 OS=Helicoverpa zea nudivirus 2 OX=1128424 GN=orf14 PE=4 SV=1
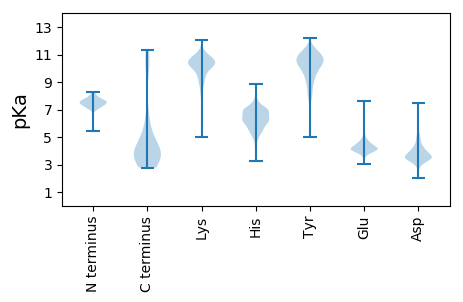
MM1 pKa = 7.69SIQGGGCGADD11 pKa = 2.83SSEE14 pKa = 5.12CNDD17 pKa = 3.69CQFDD21 pKa = 3.77KK22 pKa = 11.33AGTGEE27 pKa = 3.98NVRR30 pKa = 11.84RR31 pKa = 11.84GQSCSTDD38 pKa = 3.21AEE40 pKa = 4.29PVLGSFEE47 pKa = 4.18YY48 pKa = 10.47TDD50 pKa = 4.85CNPADD55 pKa = 4.1TIAAGNIVDD64 pKa = 4.97LKK66 pKa = 9.45NVRR69 pKa = 11.84YY70 pKa = 9.92GSINTSVSSDD80 pKa = 3.43CSEE83 pKa = 4.62DD84 pKa = 3.36SSNVYY89 pKa = 10.21SNRR92 pKa = 11.84ATISATPVVIQDD104 pKa = 3.91SEE106 pKa = 4.83DD107 pKa = 3.52EE108 pKa = 4.14QFMDD112 pKa = 5.1DD113 pKa = 3.74SKK115 pKa = 11.53SVHH118 pKa = 6.21KK119 pKa = 10.78GSFIEE124 pKa = 4.11YY125 pKa = 9.7RR126 pKa = 11.84AISKK130 pKa = 10.54NEE132 pKa = 3.74YY133 pKa = 8.89NQNEE137 pKa = 4.37YY138 pKa = 9.92NQNEE142 pKa = 4.39YY143 pKa = 9.93NQNEE147 pKa = 4.4YY148 pKa = 11.11NKK150 pKa = 10.46NEE152 pKa = 4.05YY153 pKa = 8.77NQNEE157 pKa = 4.36YY158 pKa = 11.07NKK160 pKa = 10.51NEE162 pKa = 4.05YY163 pKa = 8.68TSKK166 pKa = 10.97NDD168 pKa = 3.21YY169 pKa = 9.91NSTMRR174 pKa = 11.84RR175 pKa = 11.84CKK177 pKa = 9.41LHH179 pKa = 5.6RR180 pKa = 11.84VKK182 pKa = 10.8RR183 pKa = 11.84SIVEE187 pKa = 4.22SFEE190 pKa = 4.41STDD193 pKa = 3.03ATPNVDD199 pKa = 2.61VDD201 pKa = 3.26AMEE204 pKa = 4.98RR205 pKa = 11.84GRR207 pKa = 11.84GASAIEE213 pKa = 4.29KK214 pKa = 10.32ANTMDD219 pKa = 3.73NSNKK223 pKa = 10.43DD224 pKa = 3.48GVVEE228 pKa = 3.83EE229 pKa = 4.98SIVVVNGSTIDD240 pKa = 3.62GSIVDD245 pKa = 4.56GGTIDD250 pKa = 4.43GGIIDD255 pKa = 4.11TSNVIEE261 pKa = 4.33ASIIDD266 pKa = 3.65VDD268 pKa = 4.59SEE270 pKa = 4.39VSQVNNTNTGSGNVGVEE287 pKa = 4.07SSNVEE292 pKa = 4.16SNNVEE297 pKa = 3.85RR298 pKa = 11.84SNVEE302 pKa = 3.77TSNVDD307 pKa = 3.36VNVDD311 pKa = 3.55AEE313 pKa = 4.35SSNAEE318 pKa = 3.56VDD320 pKa = 3.32VDD322 pKa = 4.07VDD324 pKa = 4.79AEE326 pKa = 4.33YY327 pKa = 10.95TNNDD331 pKa = 3.29DD332 pKa = 3.6ATNVGEE338 pKa = 4.25TTEE341 pKa = 5.8DD342 pKa = 3.21NTEE345 pKa = 4.06YY346 pKa = 9.99NTDD349 pKa = 3.1EE350 pKa = 4.36TTNEE354 pKa = 3.89NTDD357 pKa = 3.28EE358 pKa = 4.2NTTDD362 pKa = 3.8DD363 pKa = 4.09TDD365 pKa = 3.07EE366 pKa = 4.49HH367 pKa = 6.4TDD369 pKa = 3.31EE370 pKa = 4.55NTDD373 pKa = 3.25EE374 pKa = 4.34NTNEE378 pKa = 4.04DD379 pKa = 3.42TDD381 pKa = 3.94EE382 pKa = 4.43NTDD385 pKa = 3.08EE386 pKa = 4.14TTEE389 pKa = 4.17YY390 pKa = 9.63NTEE393 pKa = 4.47DD394 pKa = 3.79YY395 pKa = 11.44NSTDD399 pKa = 3.32VNIDD403 pKa = 2.93ASRR406 pKa = 11.84IEE408 pKa = 4.42TINTSGNCFVHH419 pKa = 7.14FLLTDD424 pKa = 3.51TRR426 pKa = 11.84FLNYY430 pKa = 8.52TLSMHH435 pKa = 7.11ASLIHH440 pKa = 6.44IVYY443 pKa = 8.87TCMYY447 pKa = 9.18KK448 pKa = 10.56
MM1 pKa = 7.69SIQGGGCGADD11 pKa = 2.83SSEE14 pKa = 5.12CNDD17 pKa = 3.69CQFDD21 pKa = 3.77KK22 pKa = 11.33AGTGEE27 pKa = 3.98NVRR30 pKa = 11.84RR31 pKa = 11.84GQSCSTDD38 pKa = 3.21AEE40 pKa = 4.29PVLGSFEE47 pKa = 4.18YY48 pKa = 10.47TDD50 pKa = 4.85CNPADD55 pKa = 4.1TIAAGNIVDD64 pKa = 4.97LKK66 pKa = 9.45NVRR69 pKa = 11.84YY70 pKa = 9.92GSINTSVSSDD80 pKa = 3.43CSEE83 pKa = 4.62DD84 pKa = 3.36SSNVYY89 pKa = 10.21SNRR92 pKa = 11.84ATISATPVVIQDD104 pKa = 3.91SEE106 pKa = 4.83DD107 pKa = 3.52EE108 pKa = 4.14QFMDD112 pKa = 5.1DD113 pKa = 3.74SKK115 pKa = 11.53SVHH118 pKa = 6.21KK119 pKa = 10.78GSFIEE124 pKa = 4.11YY125 pKa = 9.7RR126 pKa = 11.84AISKK130 pKa = 10.54NEE132 pKa = 3.74YY133 pKa = 8.89NQNEE137 pKa = 4.37YY138 pKa = 9.92NQNEE142 pKa = 4.39YY143 pKa = 9.93NQNEE147 pKa = 4.4YY148 pKa = 11.11NKK150 pKa = 10.46NEE152 pKa = 4.05YY153 pKa = 8.77NQNEE157 pKa = 4.36YY158 pKa = 11.07NKK160 pKa = 10.51NEE162 pKa = 4.05YY163 pKa = 8.68TSKK166 pKa = 10.97NDD168 pKa = 3.21YY169 pKa = 9.91NSTMRR174 pKa = 11.84RR175 pKa = 11.84CKK177 pKa = 9.41LHH179 pKa = 5.6RR180 pKa = 11.84VKK182 pKa = 10.8RR183 pKa = 11.84SIVEE187 pKa = 4.22SFEE190 pKa = 4.41STDD193 pKa = 3.03ATPNVDD199 pKa = 2.61VDD201 pKa = 3.26AMEE204 pKa = 4.98RR205 pKa = 11.84GRR207 pKa = 11.84GASAIEE213 pKa = 4.29KK214 pKa = 10.32ANTMDD219 pKa = 3.73NSNKK223 pKa = 10.43DD224 pKa = 3.48GVVEE228 pKa = 3.83EE229 pKa = 4.98SIVVVNGSTIDD240 pKa = 3.62GSIVDD245 pKa = 4.56GGTIDD250 pKa = 4.43GGIIDD255 pKa = 4.11TSNVIEE261 pKa = 4.33ASIIDD266 pKa = 3.65VDD268 pKa = 4.59SEE270 pKa = 4.39VSQVNNTNTGSGNVGVEE287 pKa = 4.07SSNVEE292 pKa = 4.16SNNVEE297 pKa = 3.85RR298 pKa = 11.84SNVEE302 pKa = 3.77TSNVDD307 pKa = 3.36VNVDD311 pKa = 3.55AEE313 pKa = 4.35SSNAEE318 pKa = 3.56VDD320 pKa = 3.32VDD322 pKa = 4.07VDD324 pKa = 4.79AEE326 pKa = 4.33YY327 pKa = 10.95TNNDD331 pKa = 3.29DD332 pKa = 3.6ATNVGEE338 pKa = 4.25TTEE341 pKa = 5.8DD342 pKa = 3.21NTEE345 pKa = 4.06YY346 pKa = 9.99NTDD349 pKa = 3.1EE350 pKa = 4.36TTNEE354 pKa = 3.89NTDD357 pKa = 3.28EE358 pKa = 4.2NTTDD362 pKa = 3.8DD363 pKa = 4.09TDD365 pKa = 3.07EE366 pKa = 4.49HH367 pKa = 6.4TDD369 pKa = 3.31EE370 pKa = 4.55NTDD373 pKa = 3.25EE374 pKa = 4.34NTNEE378 pKa = 4.04DD379 pKa = 3.42TDD381 pKa = 3.94EE382 pKa = 4.43NTDD385 pKa = 3.08EE386 pKa = 4.14TTEE389 pKa = 4.17YY390 pKa = 9.63NTEE393 pKa = 4.47DD394 pKa = 3.79YY395 pKa = 11.44NSTDD399 pKa = 3.32VNIDD403 pKa = 2.93ASRR406 pKa = 11.84IEE408 pKa = 4.42TINTSGNCFVHH419 pKa = 7.14FLLTDD424 pKa = 3.51TRR426 pKa = 11.84FLNYY430 pKa = 8.52TLSMHH435 pKa = 7.11ASLIHH440 pKa = 6.44IVYY443 pKa = 8.87TCMYY447 pKa = 9.18KK448 pKa = 10.56
Molecular weight: 49.27 kDa
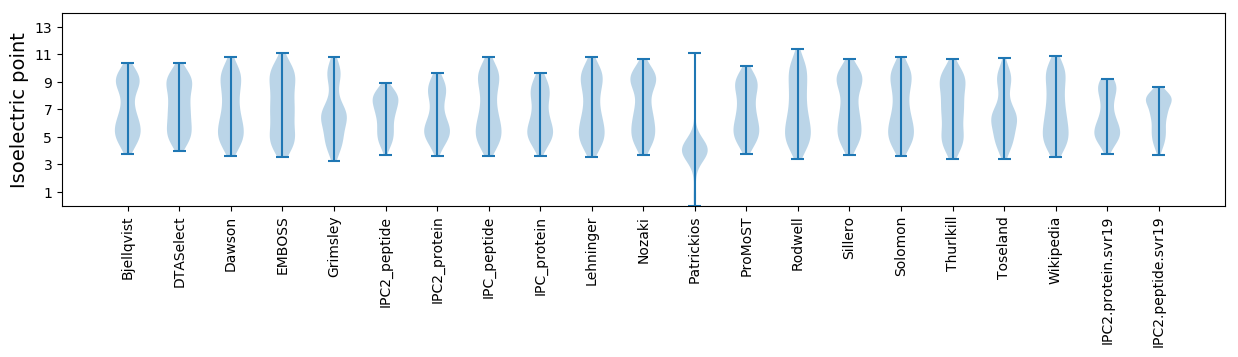
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9I0D4|G9I0D4_HZNV2 Baculovirus 38k protein OS=Helicoverpa zea nudivirus 2 OX=1128424 GN=orf108 PE=4 SV=1
MM1 pKa = 7.25QILMRR6 pKa = 11.84SSTYY10 pKa = 8.29EE11 pKa = 3.55TFMTISSFVNKK22 pKa = 10.0RR23 pKa = 11.84LFSTATMCDD32 pKa = 3.5ANLSLYY38 pKa = 10.11RR39 pKa = 11.84VALLKK44 pKa = 10.94KK45 pKa = 7.97NTASVVHH52 pKa = 6.77FIHH55 pKa = 6.14NLSYY59 pKa = 10.85TSTSNTTSTTTTTSTTSTTTHH80 pKa = 5.98HH81 pKa = 7.01RR82 pKa = 11.84GLPILVIFQSCNINN96 pKa = 3.1
MM1 pKa = 7.25QILMRR6 pKa = 11.84SSTYY10 pKa = 8.29EE11 pKa = 3.55TFMTISSFVNKK22 pKa = 10.0RR23 pKa = 11.84LFSTATMCDD32 pKa = 3.5ANLSLYY38 pKa = 10.11RR39 pKa = 11.84VALLKK44 pKa = 10.94KK45 pKa = 7.97NTASVVHH52 pKa = 6.77FIHH55 pKa = 6.14NLSYY59 pKa = 10.85TSTSNTTSTTTTTSTTSTTTHH80 pKa = 5.98HH81 pKa = 7.01RR82 pKa = 11.84GLPILVIFQSCNINN96 pKa = 3.1
Molecular weight: 10.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
52527 |
63 |
1955 |
464.8 |
52.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.612 ± 0.274 | 1.959 ± 0.146 |
6.701 ± 0.244 | 6.3 ± 0.335 |
3.621 ± 0.14 | 4.245 ± 0.154 |
2.189 ± 0.103 | 5.344 ± 0.176 |
7.014 ± 0.299 | 7.828 ± 0.297 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.61 ± 0.109 | 6.996 ± 0.404 |
4.512 ± 0.221 | 3.115 ± 0.167 |
4.636 ± 0.157 | 9.085 ± 0.374 |
7.343 ± 0.355 | 6.509 ± 0.159 |
0.52 ± 0.043 | 3.861 ± 0.199 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
