
Lachnoclostridium sp. An196
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; unclassified Lachnoclostridium
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
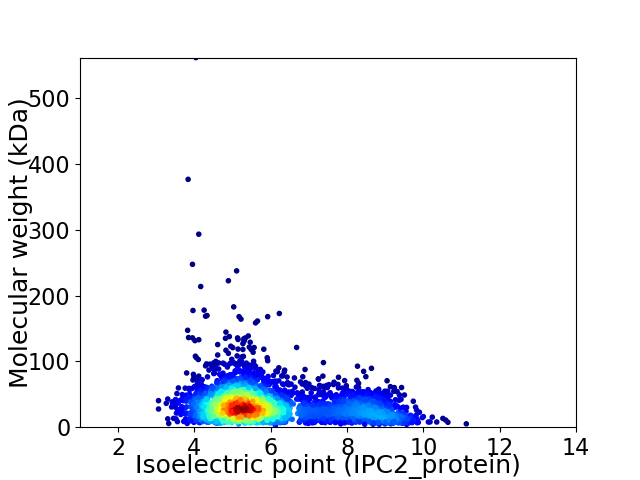
Virtual 2D-PAGE plot for 3081 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
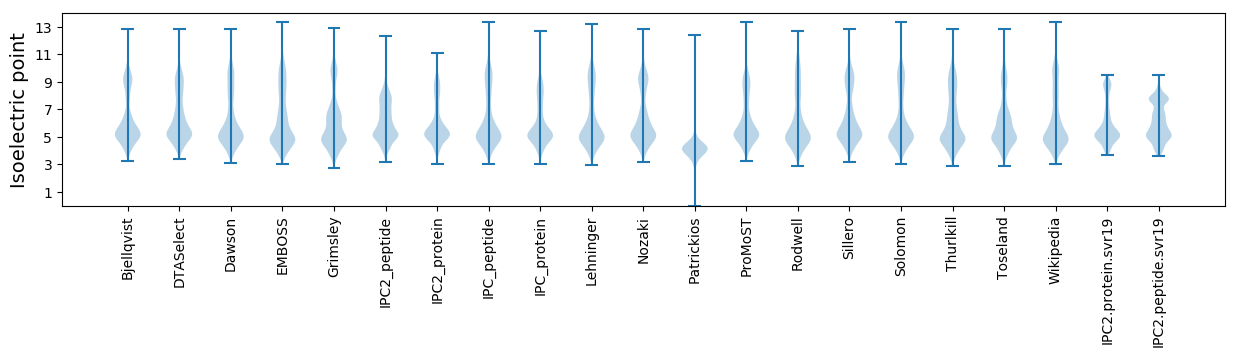
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4IPK1|A0A1Y4IPK1_9FIRM Probable cell division protein WhiA OS=Lachnoclostridium sp. An196 OX=1965583 GN=whiA PE=3 SV=1
MM1 pKa = 7.13KK2 pKa = 10.43RR3 pKa = 11.84KK4 pKa = 9.15LAVMAMAGVLAVSAVACGSSTTSTDD29 pKa = 3.38SADD32 pKa = 3.56TATEE36 pKa = 4.01STDD39 pKa = 3.28AAEE42 pKa = 4.53EE43 pKa = 4.3STDD46 pKa = 3.51AAAADD51 pKa = 4.0SASSGEE57 pKa = 4.22VANADD62 pKa = 4.03KK63 pKa = 10.82PLVWFNRR70 pKa = 11.84QPSNSSTGEE79 pKa = 3.63LDD81 pKa = 3.17MAALNFNEE89 pKa = 3.6NTYY92 pKa = 11.09YY93 pKa = 11.02VGFDD97 pKa = 3.41ANQGAEE103 pKa = 4.18LQGTMVLDD111 pKa = 3.99YY112 pKa = 10.96IEE114 pKa = 5.64ANIDD118 pKa = 3.7SIDD121 pKa = 3.8RR122 pKa = 11.84NGDD125 pKa = 3.54GIIGYY130 pKa = 9.13VLAIGDD136 pKa = 3.91IGHH139 pKa = 6.8NDD141 pKa = 3.2SIARR145 pKa = 11.84TRR147 pKa = 11.84GVRR150 pKa = 11.84SALGTGVDD158 pKa = 3.84ADD160 pKa = 3.82GSINSEE166 pKa = 3.81PVGTNTDD173 pKa = 3.2GSSSVVQDD181 pKa = 2.87GTIEE185 pKa = 4.29VNGQTYY191 pKa = 7.35TVRR194 pKa = 11.84EE195 pKa = 4.1LASQEE200 pKa = 4.09MKK202 pKa = 10.84NSAGATWDD210 pKa = 3.21AATAGNAINTWSASFGDD227 pKa = 3.84QIDD230 pKa = 3.96VVVSNNDD237 pKa = 3.14GMGMSMFNAWSKK249 pKa = 11.45DD250 pKa = 3.24NSVPTFGYY258 pKa = 10.14DD259 pKa = 3.22ANSDD263 pKa = 3.32AVAAIAEE270 pKa = 4.99GYY272 pKa = 10.39GGTISQHH279 pKa = 6.65ADD281 pKa = 2.86VQAYY285 pKa = 7.43LTLRR289 pKa = 11.84VLRR292 pKa = 11.84NALDD296 pKa = 3.9GVDD299 pKa = 3.46IDD301 pKa = 4.49TGIGTPDD308 pKa = 3.04EE309 pKa = 4.64AGNVLSSDD317 pKa = 3.51VFTYY321 pKa = 10.81NADD324 pKa = 3.32EE325 pKa = 4.15RR326 pKa = 11.84SYY328 pKa = 11.15YY329 pKa = 10.26ALNVAVTADD338 pKa = 3.58NYY340 pKa = 11.01QDD342 pKa = 4.45FLDD345 pKa = 4.02STVVYY350 pKa = 10.65APVSNQLDD358 pKa = 3.96AEE360 pKa = 4.2AHH362 pKa = 5.04PTKK365 pKa = 10.73NVWLNIYY372 pKa = 9.75NAADD376 pKa = 3.49NFLSATYY383 pKa = 10.31QPLLQNYY390 pKa = 9.65DD391 pKa = 3.59DD392 pKa = 4.97LLNLNVEE399 pKa = 4.63YY400 pKa = 10.44IGGDD404 pKa = 3.26GQTEE408 pKa = 4.46SNITNRR414 pKa = 11.84LGNPSQYY421 pKa = 11.16DD422 pKa = 3.19AFAINMVKK430 pKa = 9.6TDD432 pKa = 3.32NATSYY437 pKa = 7.53TTLLSQQ443 pKa = 4.58
MM1 pKa = 7.13KK2 pKa = 10.43RR3 pKa = 11.84KK4 pKa = 9.15LAVMAMAGVLAVSAVACGSSTTSTDD29 pKa = 3.38SADD32 pKa = 3.56TATEE36 pKa = 4.01STDD39 pKa = 3.28AAEE42 pKa = 4.53EE43 pKa = 4.3STDD46 pKa = 3.51AAAADD51 pKa = 4.0SASSGEE57 pKa = 4.22VANADD62 pKa = 4.03KK63 pKa = 10.82PLVWFNRR70 pKa = 11.84QPSNSSTGEE79 pKa = 3.63LDD81 pKa = 3.17MAALNFNEE89 pKa = 3.6NTYY92 pKa = 11.09YY93 pKa = 11.02VGFDD97 pKa = 3.41ANQGAEE103 pKa = 4.18LQGTMVLDD111 pKa = 3.99YY112 pKa = 10.96IEE114 pKa = 5.64ANIDD118 pKa = 3.7SIDD121 pKa = 3.8RR122 pKa = 11.84NGDD125 pKa = 3.54GIIGYY130 pKa = 9.13VLAIGDD136 pKa = 3.91IGHH139 pKa = 6.8NDD141 pKa = 3.2SIARR145 pKa = 11.84TRR147 pKa = 11.84GVRR150 pKa = 11.84SALGTGVDD158 pKa = 3.84ADD160 pKa = 3.82GSINSEE166 pKa = 3.81PVGTNTDD173 pKa = 3.2GSSSVVQDD181 pKa = 2.87GTIEE185 pKa = 4.29VNGQTYY191 pKa = 7.35TVRR194 pKa = 11.84EE195 pKa = 4.1LASQEE200 pKa = 4.09MKK202 pKa = 10.84NSAGATWDD210 pKa = 3.21AATAGNAINTWSASFGDD227 pKa = 3.84QIDD230 pKa = 3.96VVVSNNDD237 pKa = 3.14GMGMSMFNAWSKK249 pKa = 11.45DD250 pKa = 3.24NSVPTFGYY258 pKa = 10.14DD259 pKa = 3.22ANSDD263 pKa = 3.32AVAAIAEE270 pKa = 4.99GYY272 pKa = 10.39GGTISQHH279 pKa = 6.65ADD281 pKa = 2.86VQAYY285 pKa = 7.43LTLRR289 pKa = 11.84VLRR292 pKa = 11.84NALDD296 pKa = 3.9GVDD299 pKa = 3.46IDD301 pKa = 4.49TGIGTPDD308 pKa = 3.04EE309 pKa = 4.64AGNVLSSDD317 pKa = 3.51VFTYY321 pKa = 10.81NADD324 pKa = 3.32EE325 pKa = 4.15RR326 pKa = 11.84SYY328 pKa = 11.15YY329 pKa = 10.26ALNVAVTADD338 pKa = 3.58NYY340 pKa = 11.01QDD342 pKa = 4.45FLDD345 pKa = 4.02STVVYY350 pKa = 10.65APVSNQLDD358 pKa = 3.96AEE360 pKa = 4.2AHH362 pKa = 5.04PTKK365 pKa = 10.73NVWLNIYY372 pKa = 9.75NAADD376 pKa = 3.49NFLSATYY383 pKa = 10.31QPLLQNYY390 pKa = 9.65DD391 pKa = 3.59DD392 pKa = 4.97LLNLNVEE399 pKa = 4.63YY400 pKa = 10.44IGGDD404 pKa = 3.26GQTEE408 pKa = 4.46SNITNRR414 pKa = 11.84LGNPSQYY421 pKa = 11.16DD422 pKa = 3.19AFAINMVKK430 pKa = 9.6TDD432 pKa = 3.32NATSYY437 pKa = 7.53TTLLSQQ443 pKa = 4.58
Molecular weight: 46.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4IPP2|A0A1Y4IPP2_9FIRM ABC transporter ATP-binding protein OS=Lachnoclostridium sp. An196 OX=1965583 GN=B5F29_02840 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
977824 |
25 |
5186 |
317.4 |
35.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.996 ± 0.041 | 1.565 ± 0.021 |
5.738 ± 0.037 | 8.2 ± 0.048 |
3.885 ± 0.03 | 7.417 ± 0.043 |
1.742 ± 0.021 | 6.54 ± 0.037 |
5.708 ± 0.048 | 9.346 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.077 ± 0.029 | 3.661 ± 0.031 |
3.531 ± 0.023 | 3.402 ± 0.028 |
5.457 ± 0.046 | 5.565 ± 0.041 |
5.205 ± 0.051 | 6.848 ± 0.039 |
1.036 ± 0.016 | 4.08 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
