
Sulfurovum sp. UBA12169
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Sulfurovaceae; Sulfurovum; unclassified Sulfurovum
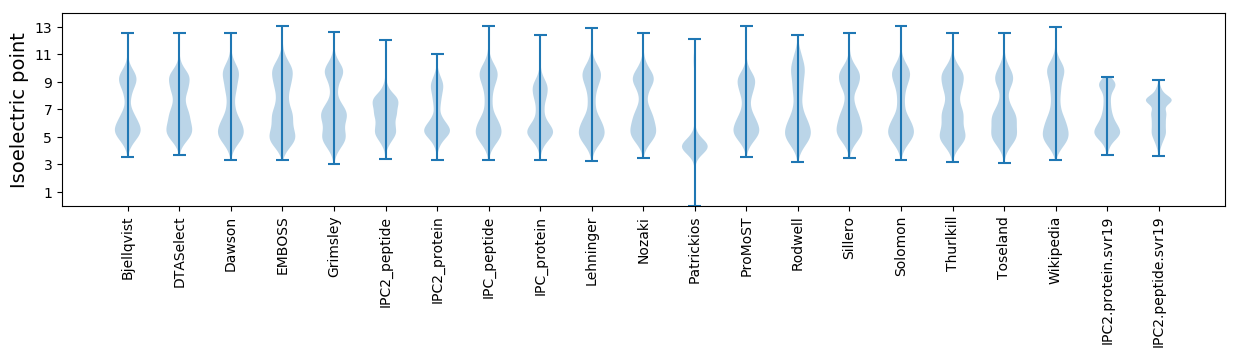
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
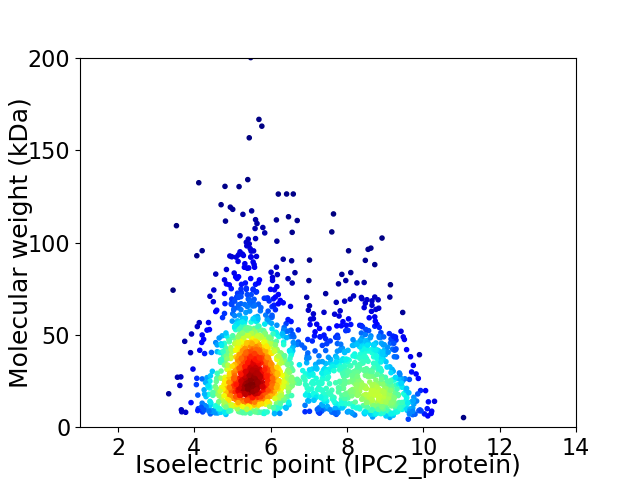
Virtual 2D-PAGE plot for 1860 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D3WS76|A0A2D3WS76_9PROT Two-component system response regulator OS=Sulfurovum sp. UBA12169 OX=2015906 GN=CFH81_07960 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 9.02KK3 pKa = 8.18TLLLSVVASTMIMAGGDD20 pKa = 3.49IAPVEE25 pKa = 4.32PVVEE29 pKa = 4.79TPAPAVSGWEE39 pKa = 4.06FSGQAALYY47 pKa = 8.58YY48 pKa = 8.49QTHH51 pKa = 7.01DD52 pKa = 3.2AFGYY56 pKa = 10.61SDD58 pKa = 6.04LFDD61 pKa = 3.99QDD63 pKa = 3.44SSAANAGLQLRR74 pKa = 11.84AVNADD79 pKa = 4.08LYY81 pKa = 10.56NGIGAGVEE89 pKa = 4.16VSGLATFDD97 pKa = 4.23LEE99 pKa = 4.26NSVVSDD105 pKa = 3.17VMQGFGTDD113 pKa = 3.89DD114 pKa = 4.92LDD116 pKa = 4.79GGWISQMYY124 pKa = 8.19LTYY127 pKa = 10.99GFDD130 pKa = 3.29NTSFKK135 pKa = 10.77LGRR138 pKa = 11.84QEE140 pKa = 4.22LPKK143 pKa = 10.51ALSPFAFSEE152 pKa = 4.03EE153 pKa = 3.96WNVFKK158 pKa = 10.37NTYY161 pKa = 9.88DD162 pKa = 3.24ALLVVNSDD170 pKa = 2.98ISNTTLVGAWVRR182 pKa = 11.84SANQNGYY189 pKa = 9.98GLGAMNGFNPSFNFQAGMPPILGTPASDD217 pKa = 3.0MNSFEE222 pKa = 5.18EE223 pKa = 4.07INGDD227 pKa = 3.25HH228 pKa = 6.43GVFMLTAQNKK238 pKa = 9.58SIEE241 pKa = 4.18NLTLTGSWYY250 pKa = 10.46YY251 pKa = 11.39SNEE254 pKa = 3.45FLTNDD259 pKa = 4.51DD260 pKa = 5.95LNILWGDD267 pKa = 3.35AQYY270 pKa = 11.77NFMQNYY276 pKa = 8.01TLGLQGGTVMHH287 pKa = 7.25DD288 pKa = 2.98AFEE291 pKa = 4.49TFNSGDD297 pKa = 3.47DD298 pKa = 3.67TVAFGAMIGGKK309 pKa = 9.88YY310 pKa = 10.27DD311 pKa = 3.29IFHH314 pKa = 6.93GALAFSSVDD323 pKa = 3.31DD324 pKa = 4.16GGFGVFNVGGQQTALYY340 pKa = 7.88TQMMFNEE347 pKa = 4.08RR348 pKa = 11.84AIAFDD353 pKa = 3.54SDD355 pKa = 3.36TFMVKK360 pKa = 10.14VGADD364 pKa = 3.25VFGGKK369 pKa = 8.76VCAAYY374 pKa = 10.29GMSDD378 pKa = 3.25MGMGDD383 pKa = 3.63YY384 pKa = 11.47NEE386 pKa = 4.96FDD388 pKa = 3.51LSYY391 pKa = 10.24SAKK394 pKa = 9.05VTDD397 pKa = 5.53SIDD400 pKa = 3.11LTAAYY405 pKa = 10.33AFMDD409 pKa = 4.16ADD411 pKa = 4.29VEE413 pKa = 5.38DD414 pKa = 5.68FDD416 pKa = 5.04DD417 pKa = 4.04TNNVIRR423 pKa = 11.84VIGRR427 pKa = 11.84YY428 pKa = 8.34NFF430 pKa = 3.39
MM1 pKa = 7.74KK2 pKa = 9.02KK3 pKa = 8.18TLLLSVVASTMIMAGGDD20 pKa = 3.49IAPVEE25 pKa = 4.32PVVEE29 pKa = 4.79TPAPAVSGWEE39 pKa = 4.06FSGQAALYY47 pKa = 8.58YY48 pKa = 8.49QTHH51 pKa = 7.01DD52 pKa = 3.2AFGYY56 pKa = 10.61SDD58 pKa = 6.04LFDD61 pKa = 3.99QDD63 pKa = 3.44SSAANAGLQLRR74 pKa = 11.84AVNADD79 pKa = 4.08LYY81 pKa = 10.56NGIGAGVEE89 pKa = 4.16VSGLATFDD97 pKa = 4.23LEE99 pKa = 4.26NSVVSDD105 pKa = 3.17VMQGFGTDD113 pKa = 3.89DD114 pKa = 4.92LDD116 pKa = 4.79GGWISQMYY124 pKa = 8.19LTYY127 pKa = 10.99GFDD130 pKa = 3.29NTSFKK135 pKa = 10.77LGRR138 pKa = 11.84QEE140 pKa = 4.22LPKK143 pKa = 10.51ALSPFAFSEE152 pKa = 4.03EE153 pKa = 3.96WNVFKK158 pKa = 10.37NTYY161 pKa = 9.88DD162 pKa = 3.24ALLVVNSDD170 pKa = 2.98ISNTTLVGAWVRR182 pKa = 11.84SANQNGYY189 pKa = 9.98GLGAMNGFNPSFNFQAGMPPILGTPASDD217 pKa = 3.0MNSFEE222 pKa = 5.18EE223 pKa = 4.07INGDD227 pKa = 3.25HH228 pKa = 6.43GVFMLTAQNKK238 pKa = 9.58SIEE241 pKa = 4.18NLTLTGSWYY250 pKa = 10.46YY251 pKa = 11.39SNEE254 pKa = 3.45FLTNDD259 pKa = 4.51DD260 pKa = 5.95LNILWGDD267 pKa = 3.35AQYY270 pKa = 11.77NFMQNYY276 pKa = 8.01TLGLQGGTVMHH287 pKa = 7.25DD288 pKa = 2.98AFEE291 pKa = 4.49TFNSGDD297 pKa = 3.47DD298 pKa = 3.67TVAFGAMIGGKK309 pKa = 9.88YY310 pKa = 10.27DD311 pKa = 3.29IFHH314 pKa = 6.93GALAFSSVDD323 pKa = 3.31DD324 pKa = 4.16GGFGVFNVGGQQTALYY340 pKa = 7.88TQMMFNEE347 pKa = 4.08RR348 pKa = 11.84AIAFDD353 pKa = 3.54SDD355 pKa = 3.36TFMVKK360 pKa = 10.14VGADD364 pKa = 3.25VFGGKK369 pKa = 8.76VCAAYY374 pKa = 10.29GMSDD378 pKa = 3.25MGMGDD383 pKa = 3.63YY384 pKa = 11.47NEE386 pKa = 4.96FDD388 pKa = 3.51LSYY391 pKa = 10.24SAKK394 pKa = 9.05VTDD397 pKa = 5.53SIDD400 pKa = 3.11LTAAYY405 pKa = 10.33AFMDD409 pKa = 4.16ADD411 pKa = 4.29VEE413 pKa = 5.38DD414 pKa = 5.68FDD416 pKa = 5.04DD417 pKa = 4.04TNNVIRR423 pKa = 11.84VIGRR427 pKa = 11.84YY428 pKa = 8.34NFF430 pKa = 3.39
Molecular weight: 46.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D3WHJ4|A0A2D3WHJ4_9PROT MFS transporter OS=Sulfurovum sp. UBA12169 OX=2015906 GN=CFH81_06545 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.39RR3 pKa = 11.84TYY5 pKa = 10.06QPHH8 pKa = 4.56TTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.24GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.87TKK25 pKa = 10.11NGRR28 pKa = 11.84KK29 pKa = 9.33VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.77KK41 pKa = 10.27LAAA44 pKa = 4.49
MM1 pKa = 7.28KK2 pKa = 9.39RR3 pKa = 11.84TYY5 pKa = 10.06QPHH8 pKa = 4.56TTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.24GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.87TKK25 pKa = 10.11NGRR28 pKa = 11.84KK29 pKa = 9.33VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.77KK41 pKa = 10.27LAAA44 pKa = 4.49
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
564002 |
37 |
1807 |
303.2 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.87 ± 0.056 | 0.939 ± 0.023 |
5.403 ± 0.042 | 7.136 ± 0.054 |
4.749 ± 0.047 | 6.551 ± 0.043 |
2.132 ± 0.025 | 7.959 ± 0.054 |
7.957 ± 0.054 | 9.607 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.819 ± 0.029 | 4.3 ± 0.04 |
3.311 ± 0.034 | 3.34 ± 0.032 |
3.791 ± 0.036 | 6.0 ± 0.042 |
5.195 ± 0.046 | 6.275 ± 0.046 |
0.844 ± 0.018 | 3.821 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
