
Rinderpest morbillivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Morbillivirus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
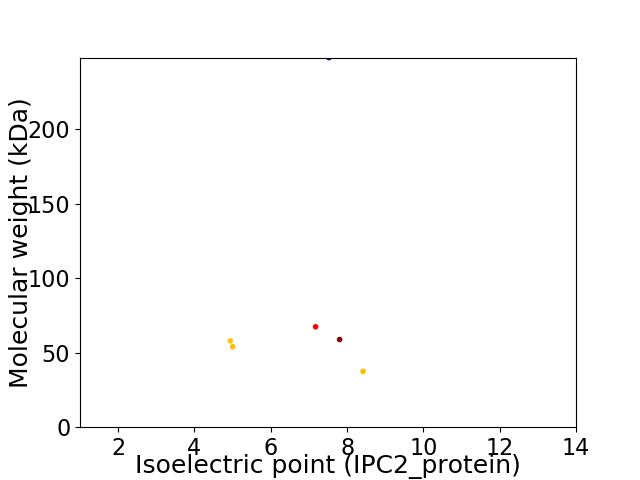
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H5BN48|A0A0H5BN48_9MONO RNA-directed RNA polymerase L OS=Rinderpest morbillivirus OX=11241 GN=L PE=3 SV=1
MM1 pKa = 7.87ASLLKK6 pKa = 10.78SLALFKK12 pKa = 10.79KK13 pKa = 10.55NKK15 pKa = 9.77DD16 pKa = 3.72KK17 pKa = 11.19PPLAAGSGGAIRR29 pKa = 11.84GIKK32 pKa = 9.67HH33 pKa = 4.91VVIVPIPGDD42 pKa = 3.45SSITTRR48 pKa = 11.84SRR50 pKa = 11.84LLDD53 pKa = 3.8CLVKK57 pKa = 10.26MVGDD61 pKa = 4.29PDD63 pKa = 3.28ISGPKK68 pKa = 8.56LTGALISILSLFVEE82 pKa = 4.97SPGQLIQRR90 pKa = 11.84ITDD93 pKa = 3.95DD94 pKa = 3.86PDD96 pKa = 3.35ISIKK100 pKa = 10.5LVEE103 pKa = 4.51VVQSDD108 pKa = 3.6KK109 pKa = 10.51TQSGLTFASRR119 pKa = 11.84GASMDD124 pKa = 4.08DD125 pKa = 3.13EE126 pKa = 5.21ADD128 pKa = 3.54RR129 pKa = 11.84YY130 pKa = 8.44FTYY133 pKa = 10.53DD134 pKa = 3.23EE135 pKa = 4.83PNGGEE140 pKa = 3.99EE141 pKa = 3.93RR142 pKa = 11.84QSYY145 pKa = 7.97WFEE148 pKa = 3.59NRR150 pKa = 11.84EE151 pKa = 3.84IQDD154 pKa = 3.55IEE156 pKa = 4.3VQDD159 pKa = 3.84PEE161 pKa = 4.65GFNMILATILAQIWILLAKK180 pKa = 10.35AVTTPDD186 pKa = 3.1TAADD190 pKa = 3.58SEE192 pKa = 4.07LRR194 pKa = 11.84RR195 pKa = 11.84WVKK198 pKa = 8.71YY199 pKa = 5.31TQQRR203 pKa = 11.84RR204 pKa = 11.84VIGEE208 pKa = 3.69FRR210 pKa = 11.84LDD212 pKa = 3.82KK213 pKa = 11.05GWLDD217 pKa = 3.56TVRR220 pKa = 11.84NRR222 pKa = 11.84IAEE225 pKa = 4.26DD226 pKa = 3.22LSLRR230 pKa = 11.84RR231 pKa = 11.84FMVALILDD239 pKa = 4.4IKK241 pKa = 9.43RR242 pKa = 11.84TPGNKK247 pKa = 8.91PRR249 pKa = 11.84IAEE252 pKa = 4.48MICDD256 pKa = 3.34IDD258 pKa = 3.96TYY260 pKa = 10.84IVEE263 pKa = 4.69AGLASFILTIKK274 pKa = 10.64FGIEE278 pKa = 3.56TMYY281 pKa = 10.26PALGLHH287 pKa = 6.29EE288 pKa = 4.47FAGEE292 pKa = 4.11LSTIEE297 pKa = 4.35SLMNLYY303 pKa = 9.91QQMGEE308 pKa = 4.06LAPYY312 pKa = 9.25MVILEE317 pKa = 4.19NSIQNKK323 pKa = 9.08FSAGAYY329 pKa = 7.48PLLWSYY335 pKa = 12.09AMGVGVEE342 pKa = 4.94LEE344 pKa = 4.14SSMGGLNFGRR354 pKa = 11.84SYY356 pKa = 11.19FDD358 pKa = 3.04PAYY361 pKa = 10.4FRR363 pKa = 11.84LGQEE367 pKa = 3.88MVRR370 pKa = 11.84RR371 pKa = 11.84SAGKK375 pKa = 9.88VSSNLASEE383 pKa = 4.65LGITEE388 pKa = 4.25EE389 pKa = 4.02EE390 pKa = 3.97AKK392 pKa = 10.57LVSEE396 pKa = 4.48IAAYY400 pKa = 9.24TGDD403 pKa = 3.9DD404 pKa = 3.6RR405 pKa = 11.84NSRR408 pKa = 11.84TSGPKK413 pKa = 7.44QTQVSFLRR421 pKa = 11.84TDD423 pKa = 3.05QGGEE427 pKa = 3.85IQHH430 pKa = 6.17NASKK434 pKa = 10.52KK435 pKa = 10.04DD436 pKa = 3.57EE437 pKa = 4.14ARR439 pKa = 11.84VPQVRR444 pKa = 11.84KK445 pKa = 7.92EE446 pKa = 4.05TWASSRR452 pKa = 11.84LDD454 pKa = 3.4RR455 pKa = 11.84YY456 pKa = 10.79KK457 pKa = 11.05EE458 pKa = 3.97DD459 pKa = 3.65TDD461 pKa = 3.91NEE463 pKa = 4.3AVSPSVKK470 pKa = 9.93TLIDD474 pKa = 3.24VDD476 pKa = 4.11TTPEE480 pKa = 4.29GDD482 pKa = 3.59TDD484 pKa = 4.05PLGSKK489 pKa = 10.16KK490 pKa = 10.31SAEE493 pKa = 4.69AILKK497 pKa = 7.94LQAMASILGDD507 pKa = 3.46STLGNDD513 pKa = 4.77SPRR516 pKa = 11.84TYY518 pKa = 10.98NDD520 pKa = 3.51KK521 pKa = 11.24DD522 pKa = 3.73LLSS525 pKa = 3.86
MM1 pKa = 7.87ASLLKK6 pKa = 10.78SLALFKK12 pKa = 10.79KK13 pKa = 10.55NKK15 pKa = 9.77DD16 pKa = 3.72KK17 pKa = 11.19PPLAAGSGGAIRR29 pKa = 11.84GIKK32 pKa = 9.67HH33 pKa = 4.91VVIVPIPGDD42 pKa = 3.45SSITTRR48 pKa = 11.84SRR50 pKa = 11.84LLDD53 pKa = 3.8CLVKK57 pKa = 10.26MVGDD61 pKa = 4.29PDD63 pKa = 3.28ISGPKK68 pKa = 8.56LTGALISILSLFVEE82 pKa = 4.97SPGQLIQRR90 pKa = 11.84ITDD93 pKa = 3.95DD94 pKa = 3.86PDD96 pKa = 3.35ISIKK100 pKa = 10.5LVEE103 pKa = 4.51VVQSDD108 pKa = 3.6KK109 pKa = 10.51TQSGLTFASRR119 pKa = 11.84GASMDD124 pKa = 4.08DD125 pKa = 3.13EE126 pKa = 5.21ADD128 pKa = 3.54RR129 pKa = 11.84YY130 pKa = 8.44FTYY133 pKa = 10.53DD134 pKa = 3.23EE135 pKa = 4.83PNGGEE140 pKa = 3.99EE141 pKa = 3.93RR142 pKa = 11.84QSYY145 pKa = 7.97WFEE148 pKa = 3.59NRR150 pKa = 11.84EE151 pKa = 3.84IQDD154 pKa = 3.55IEE156 pKa = 4.3VQDD159 pKa = 3.84PEE161 pKa = 4.65GFNMILATILAQIWILLAKK180 pKa = 10.35AVTTPDD186 pKa = 3.1TAADD190 pKa = 3.58SEE192 pKa = 4.07LRR194 pKa = 11.84RR195 pKa = 11.84WVKK198 pKa = 8.71YY199 pKa = 5.31TQQRR203 pKa = 11.84RR204 pKa = 11.84VIGEE208 pKa = 3.69FRR210 pKa = 11.84LDD212 pKa = 3.82KK213 pKa = 11.05GWLDD217 pKa = 3.56TVRR220 pKa = 11.84NRR222 pKa = 11.84IAEE225 pKa = 4.26DD226 pKa = 3.22LSLRR230 pKa = 11.84RR231 pKa = 11.84FMVALILDD239 pKa = 4.4IKK241 pKa = 9.43RR242 pKa = 11.84TPGNKK247 pKa = 8.91PRR249 pKa = 11.84IAEE252 pKa = 4.48MICDD256 pKa = 3.34IDD258 pKa = 3.96TYY260 pKa = 10.84IVEE263 pKa = 4.69AGLASFILTIKK274 pKa = 10.64FGIEE278 pKa = 3.56TMYY281 pKa = 10.26PALGLHH287 pKa = 6.29EE288 pKa = 4.47FAGEE292 pKa = 4.11LSTIEE297 pKa = 4.35SLMNLYY303 pKa = 9.91QQMGEE308 pKa = 4.06LAPYY312 pKa = 9.25MVILEE317 pKa = 4.19NSIQNKK323 pKa = 9.08FSAGAYY329 pKa = 7.48PLLWSYY335 pKa = 12.09AMGVGVEE342 pKa = 4.94LEE344 pKa = 4.14SSMGGLNFGRR354 pKa = 11.84SYY356 pKa = 11.19FDD358 pKa = 3.04PAYY361 pKa = 10.4FRR363 pKa = 11.84LGQEE367 pKa = 3.88MVRR370 pKa = 11.84RR371 pKa = 11.84SAGKK375 pKa = 9.88VSSNLASEE383 pKa = 4.65LGITEE388 pKa = 4.25EE389 pKa = 4.02EE390 pKa = 3.97AKK392 pKa = 10.57LVSEE396 pKa = 4.48IAAYY400 pKa = 9.24TGDD403 pKa = 3.9DD404 pKa = 3.6RR405 pKa = 11.84NSRR408 pKa = 11.84TSGPKK413 pKa = 7.44QTQVSFLRR421 pKa = 11.84TDD423 pKa = 3.05QGGEE427 pKa = 3.85IQHH430 pKa = 6.17NASKK434 pKa = 10.52KK435 pKa = 10.04DD436 pKa = 3.57EE437 pKa = 4.14ARR439 pKa = 11.84VPQVRR444 pKa = 11.84KK445 pKa = 7.92EE446 pKa = 4.05TWASSRR452 pKa = 11.84LDD454 pKa = 3.4RR455 pKa = 11.84YY456 pKa = 10.79KK457 pKa = 11.05EE458 pKa = 3.97DD459 pKa = 3.65TDD461 pKa = 3.91NEE463 pKa = 4.3AVSPSVKK470 pKa = 9.93TLIDD474 pKa = 3.24VDD476 pKa = 4.11TTPEE480 pKa = 4.29GDD482 pKa = 3.59TDD484 pKa = 4.05PLGSKK489 pKa = 10.16KK490 pKa = 10.31SAEE493 pKa = 4.69AILKK497 pKa = 7.94LQAMASILGDD507 pKa = 3.46STLGNDD513 pKa = 4.77SPRR516 pKa = 11.84TYY518 pKa = 10.98NDD520 pKa = 3.51KK521 pKa = 11.24DD522 pKa = 3.73LLSS525 pKa = 3.86
Molecular weight: 57.99 kDa
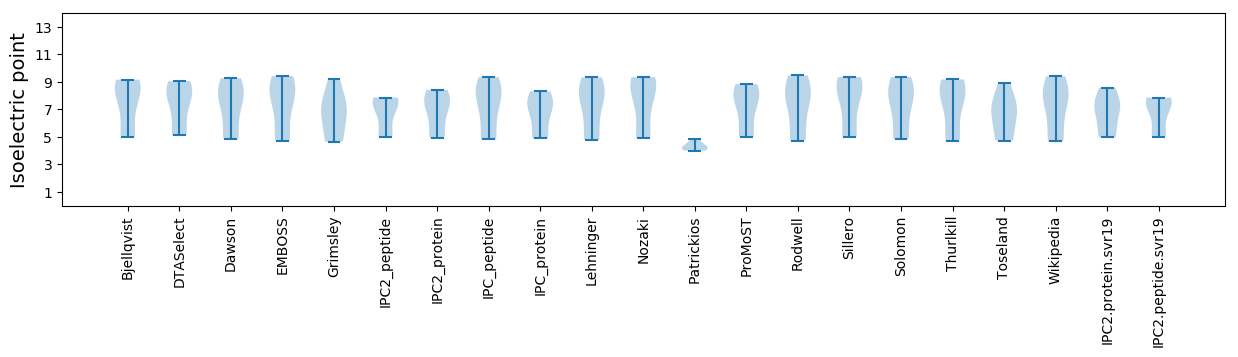
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H5B5J8|A0A0H5B5J8_9MONO Hemagglutinin glycoprotein OS=Rinderpest morbillivirus OX=11241 GN=H PE=3 SV=1
MM1 pKa = 8.04AEE3 pKa = 4.59IYY5 pKa = 10.89DD6 pKa = 3.97FDD8 pKa = 4.53KK9 pKa = 11.39SSWDD13 pKa = 3.5VKK15 pKa = 11.09GSIAPIRR22 pKa = 11.84PTTYY26 pKa = 10.35NDD28 pKa = 2.86GRR30 pKa = 11.84LIPQVRR36 pKa = 11.84VIDD39 pKa = 4.23PGLGDD44 pKa = 4.23RR45 pKa = 11.84KK46 pKa = 10.47DD47 pKa = 3.46EE48 pKa = 4.05CFMYY52 pKa = 10.31IFLLGIVEE60 pKa = 4.84DD61 pKa = 4.71SDD63 pKa = 4.01PLSPPRR69 pKa = 11.84GRR71 pKa = 11.84TFGSLPLGVGKK82 pKa = 8.51STAKK86 pKa = 10.45PEE88 pKa = 4.09EE89 pKa = 4.1LLKK92 pKa = 10.8EE93 pKa = 4.18ATDD96 pKa = 3.72LDD98 pKa = 3.63IVVRR102 pKa = 11.84RR103 pKa = 11.84TAGLNEE109 pKa = 3.91KK110 pKa = 10.39LVFYY114 pKa = 11.06NNTPLSLLTPWKK126 pKa = 10.42KK127 pKa = 10.27ILTKK131 pKa = 10.83GSVFSANQVCSAVNLIPLDD150 pKa = 3.51TPQRR154 pKa = 11.84FRR156 pKa = 11.84VVYY159 pKa = 9.03MSITRR164 pKa = 11.84LSDD167 pKa = 3.2SGCYY171 pKa = 8.94NVPRR175 pKa = 11.84KK176 pKa = 8.81MLEE179 pKa = 3.85FRR181 pKa = 11.84SANAMAFNLLVTLKK195 pKa = 9.77IDD197 pKa = 3.35TGTEE201 pKa = 3.6PGRR204 pKa = 11.84PEE206 pKa = 3.8VGAVGLSEE214 pKa = 4.15ATFMVHH220 pKa = 6.08IGNFRR225 pKa = 11.84RR226 pKa = 11.84KK227 pKa = 9.89KK228 pKa = 9.91NEE230 pKa = 3.86AYY232 pKa = 10.46SADD235 pKa = 3.68YY236 pKa = 11.12CKK238 pKa = 10.39MKK240 pKa = 10.01IEE242 pKa = 4.25KK243 pKa = 9.11MGLVFALGGIGGTSLHH259 pKa = 6.26IRR261 pKa = 11.84STGKK265 pKa = 9.26MSKK268 pKa = 8.35TLHH271 pKa = 5.95AQLGFKK277 pKa = 9.06KK278 pKa = 8.43TLCYY282 pKa = 10.37PLMDD286 pKa = 4.35INEE289 pKa = 4.46DD290 pKa = 3.64LNRR293 pKa = 11.84LLWRR297 pKa = 11.84SKK299 pKa = 10.71CKK301 pKa = 9.75IVRR304 pKa = 11.84IQAVIQPSVPQEE316 pKa = 3.3FRR318 pKa = 11.84IYY320 pKa = 11.01DD321 pKa = 3.56DD322 pKa = 5.15VIINDD327 pKa = 3.69DD328 pKa = 3.35QGLFKK333 pKa = 11.11VLL335 pKa = 3.52
MM1 pKa = 8.04AEE3 pKa = 4.59IYY5 pKa = 10.89DD6 pKa = 3.97FDD8 pKa = 4.53KK9 pKa = 11.39SSWDD13 pKa = 3.5VKK15 pKa = 11.09GSIAPIRR22 pKa = 11.84PTTYY26 pKa = 10.35NDD28 pKa = 2.86GRR30 pKa = 11.84LIPQVRR36 pKa = 11.84VIDD39 pKa = 4.23PGLGDD44 pKa = 4.23RR45 pKa = 11.84KK46 pKa = 10.47DD47 pKa = 3.46EE48 pKa = 4.05CFMYY52 pKa = 10.31IFLLGIVEE60 pKa = 4.84DD61 pKa = 4.71SDD63 pKa = 4.01PLSPPRR69 pKa = 11.84GRR71 pKa = 11.84TFGSLPLGVGKK82 pKa = 8.51STAKK86 pKa = 10.45PEE88 pKa = 4.09EE89 pKa = 4.1LLKK92 pKa = 10.8EE93 pKa = 4.18ATDD96 pKa = 3.72LDD98 pKa = 3.63IVVRR102 pKa = 11.84RR103 pKa = 11.84TAGLNEE109 pKa = 3.91KK110 pKa = 10.39LVFYY114 pKa = 11.06NNTPLSLLTPWKK126 pKa = 10.42KK127 pKa = 10.27ILTKK131 pKa = 10.83GSVFSANQVCSAVNLIPLDD150 pKa = 3.51TPQRR154 pKa = 11.84FRR156 pKa = 11.84VVYY159 pKa = 9.03MSITRR164 pKa = 11.84LSDD167 pKa = 3.2SGCYY171 pKa = 8.94NVPRR175 pKa = 11.84KK176 pKa = 8.81MLEE179 pKa = 3.85FRR181 pKa = 11.84SANAMAFNLLVTLKK195 pKa = 9.77IDD197 pKa = 3.35TGTEE201 pKa = 3.6PGRR204 pKa = 11.84PEE206 pKa = 3.8VGAVGLSEE214 pKa = 4.15ATFMVHH220 pKa = 6.08IGNFRR225 pKa = 11.84RR226 pKa = 11.84KK227 pKa = 9.89KK228 pKa = 9.91NEE230 pKa = 3.86AYY232 pKa = 10.46SADD235 pKa = 3.68YY236 pKa = 11.12CKK238 pKa = 10.39MKK240 pKa = 10.01IEE242 pKa = 4.25KK243 pKa = 9.11MGLVFALGGIGGTSLHH259 pKa = 6.26IRR261 pKa = 11.84STGKK265 pKa = 9.26MSKK268 pKa = 8.35TLHH271 pKa = 5.95AQLGFKK277 pKa = 9.06KK278 pKa = 8.43TLCYY282 pKa = 10.37PLMDD286 pKa = 4.35INEE289 pKa = 4.46DD290 pKa = 3.64LNRR293 pKa = 11.84LLWRR297 pKa = 11.84SKK299 pKa = 10.71CKK301 pKa = 9.75IVRR304 pKa = 11.84IQAVIQPSVPQEE316 pKa = 3.3FRR318 pKa = 11.84IYY320 pKa = 11.01DD321 pKa = 3.56DD322 pKa = 5.15VIINDD327 pKa = 3.69DD328 pKa = 3.35QGLFKK333 pKa = 11.11VLL335 pKa = 3.52
Molecular weight: 37.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4705 |
335 |
2183 |
784.2 |
87.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.1 ± 0.497 | 1.679 ± 0.225 |
5.845 ± 0.487 | 5.292 ± 0.329 |
3.103 ± 0.487 | 6.652 ± 0.724 |
2.232 ± 0.51 | 7.736 ± 0.219 |
5.547 ± 0.435 | 10.797 ± 0.633 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.147 ± 0.148 | 3.953 ± 0.264 |
4.612 ± 0.465 | 3.337 ± 0.195 |
5.59 ± 0.453 | 8.714 ± 0.526 |
6.015 ± 0.247 | 6.036 ± 0.226 |
1.02 ± 0.09 | 3.592 ± 0.496 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
