
Nesiotobacter exalbescens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Nesiotobacter
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
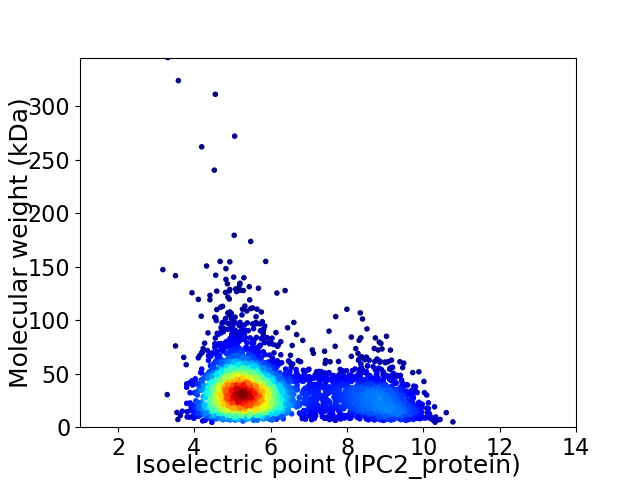
Virtual 2D-PAGE plot for 3634 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
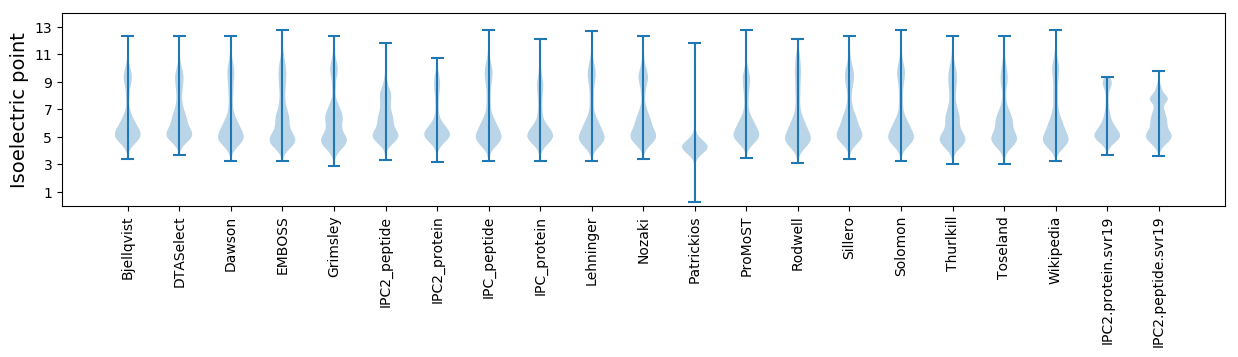
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7JF22|A0A1U7JF22_9RHOB ABC transporter domain-containing protein OS=Nesiotobacter exalbescens OX=197461 GN=A3843_14080 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 10.58LKK4 pKa = 10.83ALLLATAGAAVATSVSAADD23 pKa = 3.89LPVVAEE29 pKa = 4.07PVDD32 pKa = 3.84YY33 pKa = 11.35VQVCDD38 pKa = 4.02AYY40 pKa = 10.52GAGFFKK46 pKa = 10.7IPGKK50 pKa = 8.57DD51 pKa = 2.93TCLKK55 pKa = 9.04IGGRR59 pKa = 11.84IRR61 pKa = 11.84TQIVSDD67 pKa = 4.29NLDD70 pKa = 3.06SDD72 pKa = 4.88ADD74 pKa = 3.87YY75 pKa = 11.59DD76 pKa = 4.69DD77 pKa = 4.26YY78 pKa = 11.8SAYY81 pKa = 10.4ARR83 pKa = 11.84GYY85 pKa = 10.86LYY87 pKa = 9.46LTSMTDD93 pKa = 3.3TEE95 pKa = 4.12IGLIKK100 pKa = 9.96TYY102 pKa = 10.27TEE104 pKa = 5.68FNSEE108 pKa = 3.86WNQDD112 pKa = 3.62GGDD115 pKa = 3.7GSVDD119 pKa = 3.19VGDD122 pKa = 5.12AYY124 pKa = 11.33LQIGTNFADD133 pKa = 4.14VIIGRR138 pKa = 11.84TTSLFDD144 pKa = 3.65GFTGAVDD151 pKa = 3.23VGVIGRR157 pKa = 11.84NWSDD161 pKa = 2.95QGTLQFALTKK171 pKa = 10.58SLGNGVTAAVSAEE184 pKa = 3.82DD185 pKa = 3.13SSYY188 pKa = 11.53RR189 pKa = 11.84DD190 pKa = 3.17GDD192 pKa = 4.11TNSADD197 pKa = 3.76YY198 pKa = 10.89VGKK201 pKa = 10.64LGLAQGWGSVEE212 pKa = 3.9VAAALHH218 pKa = 6.04DD219 pKa = 4.32TVAGAVDD226 pKa = 3.4NYY228 pKa = 10.43GYY230 pKa = 11.01AVRR233 pKa = 11.84GTATVNLDD241 pKa = 3.14MLAAGDD247 pKa = 3.89AFVFQAAYY255 pKa = 10.51ADD257 pKa = 3.79AASAYY262 pKa = 9.74LGKK265 pKa = 8.88STAAWDD271 pKa = 3.9LAGDD275 pKa = 3.99EE276 pKa = 4.4AWALGAGFTHH286 pKa = 6.73YY287 pKa = 7.53WTSEE291 pKa = 3.41IATSIDD297 pKa = 3.05GSYY300 pKa = 10.79LSNDD304 pKa = 3.37DD305 pKa = 3.91ADD307 pKa = 3.95YY308 pKa = 11.42DD309 pKa = 3.38RR310 pKa = 11.84WAIDD314 pKa = 4.23GSVGWYY320 pKa = 8.06PVSGMVVALDD330 pKa = 3.52GGFASEE336 pKa = 4.42DD337 pKa = 3.59AAGVEE342 pKa = 4.02ADD344 pKa = 3.71EE345 pKa = 4.68AKK347 pKa = 10.4IGARR351 pKa = 11.84VQYY354 pKa = 9.48TFF356 pKa = 4.12
MM1 pKa = 7.52KK2 pKa = 10.58LKK4 pKa = 10.83ALLLATAGAAVATSVSAADD23 pKa = 3.89LPVVAEE29 pKa = 4.07PVDD32 pKa = 3.84YY33 pKa = 11.35VQVCDD38 pKa = 4.02AYY40 pKa = 10.52GAGFFKK46 pKa = 10.7IPGKK50 pKa = 8.57DD51 pKa = 2.93TCLKK55 pKa = 9.04IGGRR59 pKa = 11.84IRR61 pKa = 11.84TQIVSDD67 pKa = 4.29NLDD70 pKa = 3.06SDD72 pKa = 4.88ADD74 pKa = 3.87YY75 pKa = 11.59DD76 pKa = 4.69DD77 pKa = 4.26YY78 pKa = 11.8SAYY81 pKa = 10.4ARR83 pKa = 11.84GYY85 pKa = 10.86LYY87 pKa = 9.46LTSMTDD93 pKa = 3.3TEE95 pKa = 4.12IGLIKK100 pKa = 9.96TYY102 pKa = 10.27TEE104 pKa = 5.68FNSEE108 pKa = 3.86WNQDD112 pKa = 3.62GGDD115 pKa = 3.7GSVDD119 pKa = 3.19VGDD122 pKa = 5.12AYY124 pKa = 11.33LQIGTNFADD133 pKa = 4.14VIIGRR138 pKa = 11.84TTSLFDD144 pKa = 3.65GFTGAVDD151 pKa = 3.23VGVIGRR157 pKa = 11.84NWSDD161 pKa = 2.95QGTLQFALTKK171 pKa = 10.58SLGNGVTAAVSAEE184 pKa = 3.82DD185 pKa = 3.13SSYY188 pKa = 11.53RR189 pKa = 11.84DD190 pKa = 3.17GDD192 pKa = 4.11TNSADD197 pKa = 3.76YY198 pKa = 10.89VGKK201 pKa = 10.64LGLAQGWGSVEE212 pKa = 3.9VAAALHH218 pKa = 6.04DD219 pKa = 4.32TVAGAVDD226 pKa = 3.4NYY228 pKa = 10.43GYY230 pKa = 11.01AVRR233 pKa = 11.84GTATVNLDD241 pKa = 3.14MLAAGDD247 pKa = 3.89AFVFQAAYY255 pKa = 10.51ADD257 pKa = 3.79AASAYY262 pKa = 9.74LGKK265 pKa = 8.88STAAWDD271 pKa = 3.9LAGDD275 pKa = 3.99EE276 pKa = 4.4AWALGAGFTHH286 pKa = 6.73YY287 pKa = 7.53WTSEE291 pKa = 3.41IATSIDD297 pKa = 3.05GSYY300 pKa = 10.79LSNDD304 pKa = 3.37DD305 pKa = 3.91ADD307 pKa = 3.95YY308 pKa = 11.42DD309 pKa = 3.38RR310 pKa = 11.84WAIDD314 pKa = 4.23GSVGWYY320 pKa = 8.06PVSGMVVALDD330 pKa = 3.52GGFASEE336 pKa = 4.42DD337 pKa = 3.59AAGVEE342 pKa = 4.02ADD344 pKa = 3.71EE345 pKa = 4.68AKK347 pKa = 10.4IGARR351 pKa = 11.84VQYY354 pKa = 9.48TFF356 pKa = 4.12
Molecular weight: 37.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7JK17|A0A1U7JK17_9RHOB UPF0229 protein A3843_04610 OS=Nesiotobacter exalbescens OX=197461 GN=A3843_04610 PE=3 SV=1
MM1 pKa = 7.13QAPKK5 pKa = 10.1FFPGSGFPTSSVIRR19 pKa = 11.84LAARR23 pKa = 11.84YY24 pKa = 9.15RR25 pKa = 11.84EE26 pKa = 4.19AGVFPTYY33 pKa = 10.92DD34 pKa = 3.46PAVLMMALRR43 pKa = 11.84MPEE46 pKa = 4.06RR47 pKa = 11.84APEE50 pKa = 4.13ALLAEE55 pKa = 4.35ARR57 pKa = 11.84HH58 pKa = 5.69INRR61 pKa = 11.84TGRR64 pKa = 11.84WQNEE68 pKa = 3.41EE69 pKa = 3.29EE70 pKa = 4.13ARR72 pKa = 11.84RR73 pKa = 11.84KK74 pKa = 9.18RR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.45ASPRR81 pKa = 11.84KK82 pKa = 8.16PSVKK86 pKa = 10.24ARR88 pKa = 11.84VEE90 pKa = 4.19KK91 pKa = 10.52ILQQEE96 pKa = 3.94AAEE99 pKa = 4.38AGISVEE105 pKa = 4.12RR106 pKa = 11.84LRR108 pKa = 11.84GRR110 pKa = 11.84CRR112 pKa = 11.84KK113 pKa = 9.82RR114 pKa = 11.84PSLQARR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 4.14AVARR126 pKa = 11.84CHH128 pKa = 7.25DD129 pKa = 4.2EE130 pKa = 5.26LGLTCKK136 pKa = 10.57QIGEE140 pKa = 4.34YY141 pKa = 10.51FSGRR145 pKa = 11.84GAEE148 pKa = 4.09AVRR151 pKa = 11.84TLLHH155 pKa = 6.56RR156 pKa = 11.84AQAARR161 pKa = 11.84QQQAAEE167 pKa = 4.33DD168 pKa = 3.95DD169 pKa = 3.96VHH171 pKa = 8.76RR172 pKa = 11.84LMEE175 pKa = 4.32MLTEE179 pKa = 4.52AEE181 pKa = 4.21RR182 pKa = 11.84KK183 pKa = 9.42SLKK186 pKa = 9.86QRR188 pKa = 11.84ALALTIAEE196 pKa = 4.62EE197 pKa = 4.31IACKK201 pKa = 10.25HH202 pKa = 5.46EE203 pKa = 4.1RR204 pKa = 11.84TLAEE208 pKa = 3.17IRR210 pKa = 11.84ARR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84APALHH219 pKa = 5.79GVRR222 pKa = 11.84RR223 pKa = 11.84EE224 pKa = 3.83ALYY227 pKa = 9.44WAWEE231 pKa = 4.08RR232 pKa = 11.84TNLGWSEE239 pKa = 3.89LGQLFGRR246 pKa = 11.84GAGHH250 pKa = 5.8VAGLVRR256 pKa = 11.84EE257 pKa = 4.13YY258 pKa = 11.56AEE260 pKa = 4.12LHH262 pKa = 6.09GLRR265 pKa = 11.84APEE268 pKa = 3.99GMGRR272 pKa = 11.84VAAA275 pKa = 4.82
MM1 pKa = 7.13QAPKK5 pKa = 10.1FFPGSGFPTSSVIRR19 pKa = 11.84LAARR23 pKa = 11.84YY24 pKa = 9.15RR25 pKa = 11.84EE26 pKa = 4.19AGVFPTYY33 pKa = 10.92DD34 pKa = 3.46PAVLMMALRR43 pKa = 11.84MPEE46 pKa = 4.06RR47 pKa = 11.84APEE50 pKa = 4.13ALLAEE55 pKa = 4.35ARR57 pKa = 11.84HH58 pKa = 5.69INRR61 pKa = 11.84TGRR64 pKa = 11.84WQNEE68 pKa = 3.41EE69 pKa = 3.29EE70 pKa = 4.13ARR72 pKa = 11.84RR73 pKa = 11.84KK74 pKa = 9.18RR75 pKa = 11.84RR76 pKa = 11.84KK77 pKa = 9.45ASPRR81 pKa = 11.84KK82 pKa = 8.16PSVKK86 pKa = 10.24ARR88 pKa = 11.84VEE90 pKa = 4.19KK91 pKa = 10.52ILQQEE96 pKa = 3.94AAEE99 pKa = 4.38AGISVEE105 pKa = 4.12RR106 pKa = 11.84LRR108 pKa = 11.84GRR110 pKa = 11.84CRR112 pKa = 11.84KK113 pKa = 9.82RR114 pKa = 11.84PSLQARR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 4.14AVARR126 pKa = 11.84CHH128 pKa = 7.25DD129 pKa = 4.2EE130 pKa = 5.26LGLTCKK136 pKa = 10.57QIGEE140 pKa = 4.34YY141 pKa = 10.51FSGRR145 pKa = 11.84GAEE148 pKa = 4.09AVRR151 pKa = 11.84TLLHH155 pKa = 6.56RR156 pKa = 11.84AQAARR161 pKa = 11.84QQQAAEE167 pKa = 4.33DD168 pKa = 3.95DD169 pKa = 3.96VHH171 pKa = 8.76RR172 pKa = 11.84LMEE175 pKa = 4.32MLTEE179 pKa = 4.52AEE181 pKa = 4.21RR182 pKa = 11.84KK183 pKa = 9.42SLKK186 pKa = 9.86QRR188 pKa = 11.84ALALTIAEE196 pKa = 4.62EE197 pKa = 4.31IACKK201 pKa = 10.25HH202 pKa = 5.46EE203 pKa = 4.1RR204 pKa = 11.84TLAEE208 pKa = 3.17IRR210 pKa = 11.84ARR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84APALHH219 pKa = 5.79GVRR222 pKa = 11.84RR223 pKa = 11.84EE224 pKa = 3.83ALYY227 pKa = 9.44WAWEE231 pKa = 4.08RR232 pKa = 11.84TNLGWSEE239 pKa = 3.89LGQLFGRR246 pKa = 11.84GAGHH250 pKa = 5.8VAGLVRR256 pKa = 11.84EE257 pKa = 4.13YY258 pKa = 11.56AEE260 pKa = 4.12LHH262 pKa = 6.09GLRR265 pKa = 11.84APEE268 pKa = 3.99GMGRR272 pKa = 11.84VAAA275 pKa = 4.82
Molecular weight: 31.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1186005 |
38 |
3381 |
326.4 |
35.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.905 ± 0.048 | 0.917 ± 0.012 |
5.7 ± 0.043 | 6.355 ± 0.039 |
3.914 ± 0.03 | 8.07 ± 0.042 |
2.079 ± 0.019 | 5.401 ± 0.028 |
3.856 ± 0.034 | 10.195 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.642 ± 0.023 | 3.062 ± 0.024 |
4.717 ± 0.027 | 3.621 ± 0.025 |
6.045 ± 0.038 | 5.847 ± 0.032 |
5.552 ± 0.029 | 7.463 ± 0.036 |
1.217 ± 0.017 | 2.442 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
