
Desulfacinum hydrothermale DSM 13146
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Syntrophobacterales; Syntrophobacteraceae; Desulfacinum; Desulfacinum hydrothermale
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
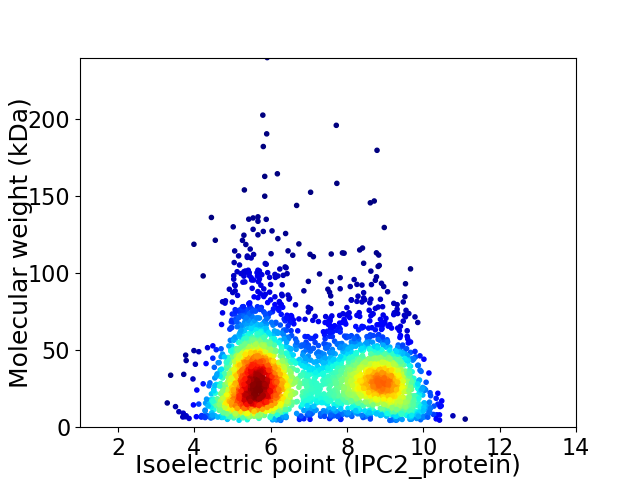
Virtual 2D-PAGE plot for 3262 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
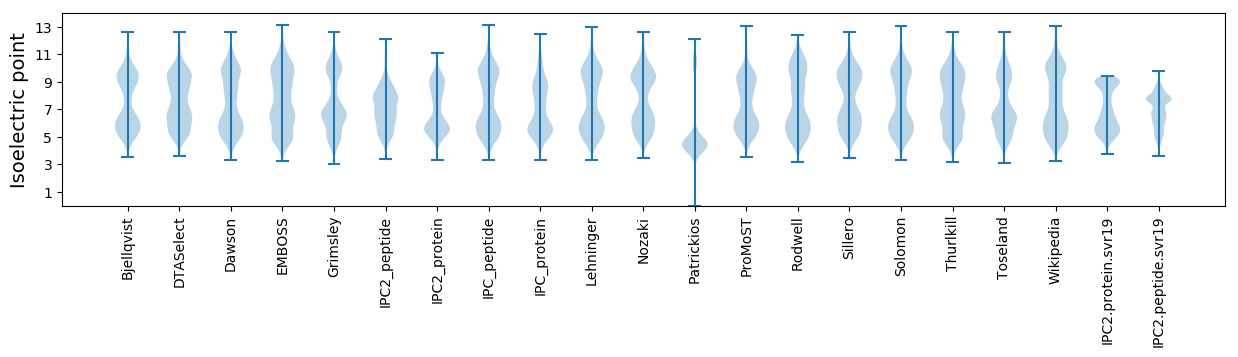
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W1X0J0|A0A1W1X0J0_9DELT Uncharacterized protein OS=Desulfacinum hydrothermale DSM 13146 OX=1121390 GN=SAMN02746041_00270 PE=4 SV=1
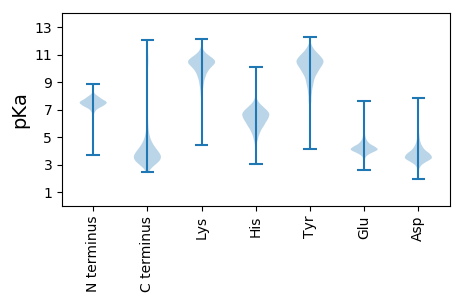
MM1 pKa = 7.81RR2 pKa = 11.84KK3 pKa = 9.32NLVVLLAVLMAVAIALPAFAVDD25 pKa = 4.18FKK27 pKa = 10.63YY28 pKa = 11.02GGMMRR33 pKa = 11.84VRR35 pKa = 11.84WYY37 pKa = 10.73SYY39 pKa = 11.51NNFDD43 pKa = 4.46GTDD46 pKa = 3.54DD47 pKa = 4.83ADD49 pKa = 3.54DD50 pKa = 3.88HH51 pKa = 7.25ANYY54 pKa = 10.02FDD56 pKa = 3.33QRR58 pKa = 11.84VRR60 pKa = 11.84MYY62 pKa = 8.27FTFVASEE69 pKa = 3.81NLQLVTKK76 pKa = 10.11WEE78 pKa = 4.03EE79 pKa = 4.08DD80 pKa = 3.81NVWGAKK86 pKa = 10.26GGDD89 pKa = 3.39RR90 pKa = 11.84DD91 pKa = 3.98LTHH94 pKa = 6.61ATSDD98 pKa = 3.41FRR100 pKa = 11.84DD101 pKa = 4.25PYY103 pKa = 11.82DD104 pKa = 3.48MEE106 pKa = 4.54TKK108 pKa = 10.38NVYY111 pKa = 10.74LDD113 pKa = 3.9FNIPNTPIRR122 pKa = 11.84AKK124 pKa = 10.51LGQQGFGLFGGRR136 pKa = 11.84IIAEE140 pKa = 4.7DD141 pKa = 3.41YY142 pKa = 10.32TGAKK146 pKa = 10.06FSMPFDD152 pKa = 3.84PVKK155 pKa = 10.45VTLGYY160 pKa = 9.99IAEE163 pKa = 4.22QNVDD167 pKa = 3.46YY168 pKa = 11.09SSEE171 pKa = 3.84KK172 pKa = 11.05DD173 pKa = 3.57NVDD176 pKa = 2.52IYY178 pKa = 11.43YY179 pKa = 10.48LQLQYY184 pKa = 11.85ANGPFSGEE192 pKa = 4.39LIGLYY197 pKa = 9.94QDD199 pKa = 4.34GSDD202 pKa = 3.38TAVSLKK208 pKa = 9.91TEE210 pKa = 4.12YY211 pKa = 11.06ADD213 pKa = 4.5PGVDD217 pKa = 3.07AQGRR221 pKa = 11.84EE222 pKa = 4.22TYY224 pKa = 10.42DD225 pKa = 3.08GYY227 pKa = 9.68TFEE230 pKa = 5.94DD231 pKa = 3.05NHH233 pKa = 7.91LFDD236 pKa = 5.97LGVKK240 pKa = 9.85LAYY243 pKa = 10.34SSDD246 pKa = 3.35MFKK249 pKa = 10.48ATVHH253 pKa = 6.49FIKK256 pKa = 10.83NFGSVDD262 pKa = 4.05LNGEE266 pKa = 3.99SADD269 pKa = 3.69YY270 pKa = 10.82KK271 pKa = 11.08GWFLGADD278 pKa = 3.26VAIPVNDD285 pKa = 3.61FTFGLGGFYY294 pKa = 7.96TTGQDD299 pKa = 3.34EE300 pKa = 4.6GDD302 pKa = 4.33DD303 pKa = 3.83DD304 pKa = 4.8VEE306 pKa = 5.03GYY308 pKa = 10.29VYY310 pKa = 10.43PGAVGDD316 pKa = 3.85SFYY319 pKa = 10.25WAEE322 pKa = 3.68IMGFGVLDD330 pKa = 4.13DD331 pKa = 4.04MTPRR335 pKa = 11.84AYY337 pKa = 9.29TGYY340 pKa = 9.22PGAGEE345 pKa = 4.0YY346 pKa = 8.71TAGGNPSNLYY356 pKa = 8.99TLYY359 pKa = 11.2ASVDD363 pKa = 3.42WQALEE368 pKa = 3.79ATKK371 pKa = 10.78LHH373 pKa = 6.11LAYY376 pKa = 10.3YY377 pKa = 9.72YY378 pKa = 10.69LGTAEE383 pKa = 5.03DD384 pKa = 4.19VVANAATGEE393 pKa = 4.16KK394 pKa = 9.69DD395 pKa = 3.38DD396 pKa = 4.38SVGSEE401 pKa = 3.49VDD403 pKa = 3.96FRR405 pKa = 11.84LTQKK409 pKa = 10.38VVDD412 pKa = 4.32GLNLDD417 pKa = 3.83VIGAYY422 pKa = 10.49LFADD426 pKa = 4.04DD427 pKa = 5.34AFSGTKK433 pKa = 10.29DD434 pKa = 3.71DD435 pKa = 4.34DD436 pKa = 3.85DD437 pKa = 6.18AYY439 pKa = 10.59EE440 pKa = 4.17VGARR444 pKa = 11.84LQWAFF449 pKa = 3.54
MM1 pKa = 7.81RR2 pKa = 11.84KK3 pKa = 9.32NLVVLLAVLMAVAIALPAFAVDD25 pKa = 4.18FKK27 pKa = 10.63YY28 pKa = 11.02GGMMRR33 pKa = 11.84VRR35 pKa = 11.84WYY37 pKa = 10.73SYY39 pKa = 11.51NNFDD43 pKa = 4.46GTDD46 pKa = 3.54DD47 pKa = 4.83ADD49 pKa = 3.54DD50 pKa = 3.88HH51 pKa = 7.25ANYY54 pKa = 10.02FDD56 pKa = 3.33QRR58 pKa = 11.84VRR60 pKa = 11.84MYY62 pKa = 8.27FTFVASEE69 pKa = 3.81NLQLVTKK76 pKa = 10.11WEE78 pKa = 4.03EE79 pKa = 4.08DD80 pKa = 3.81NVWGAKK86 pKa = 10.26GGDD89 pKa = 3.39RR90 pKa = 11.84DD91 pKa = 3.98LTHH94 pKa = 6.61ATSDD98 pKa = 3.41FRR100 pKa = 11.84DD101 pKa = 4.25PYY103 pKa = 11.82DD104 pKa = 3.48MEE106 pKa = 4.54TKK108 pKa = 10.38NVYY111 pKa = 10.74LDD113 pKa = 3.9FNIPNTPIRR122 pKa = 11.84AKK124 pKa = 10.51LGQQGFGLFGGRR136 pKa = 11.84IIAEE140 pKa = 4.7DD141 pKa = 3.41YY142 pKa = 10.32TGAKK146 pKa = 10.06FSMPFDD152 pKa = 3.84PVKK155 pKa = 10.45VTLGYY160 pKa = 9.99IAEE163 pKa = 4.22QNVDD167 pKa = 3.46YY168 pKa = 11.09SSEE171 pKa = 3.84KK172 pKa = 11.05DD173 pKa = 3.57NVDD176 pKa = 2.52IYY178 pKa = 11.43YY179 pKa = 10.48LQLQYY184 pKa = 11.85ANGPFSGEE192 pKa = 4.39LIGLYY197 pKa = 9.94QDD199 pKa = 4.34GSDD202 pKa = 3.38TAVSLKK208 pKa = 9.91TEE210 pKa = 4.12YY211 pKa = 11.06ADD213 pKa = 4.5PGVDD217 pKa = 3.07AQGRR221 pKa = 11.84EE222 pKa = 4.22TYY224 pKa = 10.42DD225 pKa = 3.08GYY227 pKa = 9.68TFEE230 pKa = 5.94DD231 pKa = 3.05NHH233 pKa = 7.91LFDD236 pKa = 5.97LGVKK240 pKa = 9.85LAYY243 pKa = 10.34SSDD246 pKa = 3.35MFKK249 pKa = 10.48ATVHH253 pKa = 6.49FIKK256 pKa = 10.83NFGSVDD262 pKa = 4.05LNGEE266 pKa = 3.99SADD269 pKa = 3.69YY270 pKa = 10.82KK271 pKa = 11.08GWFLGADD278 pKa = 3.26VAIPVNDD285 pKa = 3.61FTFGLGGFYY294 pKa = 7.96TTGQDD299 pKa = 3.34EE300 pKa = 4.6GDD302 pKa = 4.33DD303 pKa = 3.83DD304 pKa = 4.8VEE306 pKa = 5.03GYY308 pKa = 10.29VYY310 pKa = 10.43PGAVGDD316 pKa = 3.85SFYY319 pKa = 10.25WAEE322 pKa = 3.68IMGFGVLDD330 pKa = 4.13DD331 pKa = 4.04MTPRR335 pKa = 11.84AYY337 pKa = 9.29TGYY340 pKa = 9.22PGAGEE345 pKa = 4.0YY346 pKa = 8.71TAGGNPSNLYY356 pKa = 8.99TLYY359 pKa = 11.2ASVDD363 pKa = 3.42WQALEE368 pKa = 3.79ATKK371 pKa = 10.78LHH373 pKa = 6.11LAYY376 pKa = 10.3YY377 pKa = 9.72YY378 pKa = 10.69LGTAEE383 pKa = 5.03DD384 pKa = 4.19VVANAATGEE393 pKa = 4.16KK394 pKa = 9.69DD395 pKa = 3.38DD396 pKa = 4.38SVGSEE401 pKa = 3.49VDD403 pKa = 3.96FRR405 pKa = 11.84LTQKK409 pKa = 10.38VVDD412 pKa = 4.32GLNLDD417 pKa = 3.83VIGAYY422 pKa = 10.49LFADD426 pKa = 4.04DD427 pKa = 5.34AFSGTKK433 pKa = 10.29DD434 pKa = 3.71DD435 pKa = 4.34DD436 pKa = 3.85DD437 pKa = 6.18AYY439 pKa = 10.59EE440 pKa = 4.17VGARR444 pKa = 11.84LQWAFF449 pKa = 3.54
Molecular weight: 49.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W1XIP4|A0A1W1XIP4_9DELT Magnesium chelatase family protein OS=Desulfacinum hydrothermale DSM 13146 OX=1121390 GN=SAMN02746041_01807 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.19QPSNIKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 7.89RR14 pKa = 11.84THH16 pKa = 5.96GFLVRR21 pKa = 11.84MSTKK25 pKa = 10.41AGRR28 pKa = 11.84QVIKK32 pKa = 10.42RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.19QPSNIKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 7.89RR14 pKa = 11.84THH16 pKa = 5.96GFLVRR21 pKa = 11.84MSTKK25 pKa = 10.41AGRR28 pKa = 11.84QVIKK32 pKa = 10.42RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.66RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1058542 |
39 |
2151 |
324.5 |
36.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.607 ± 0.038 | 1.347 ± 0.018 |
5.239 ± 0.028 | 6.699 ± 0.041 |
3.805 ± 0.028 | 7.878 ± 0.036 |
2.424 ± 0.019 | 4.839 ± 0.032 |
4.101 ± 0.035 | 10.862 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.395 ± 0.02 | 2.387 ± 0.021 |
5.295 ± 0.025 | 3.628 ± 0.025 |
7.81 ± 0.047 | 5.038 ± 0.028 |
4.67 ± 0.024 | 8.027 ± 0.037 |
1.369 ± 0.019 | 2.579 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
