
Sphingomonadales bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; unclassified Sphingomonadales
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
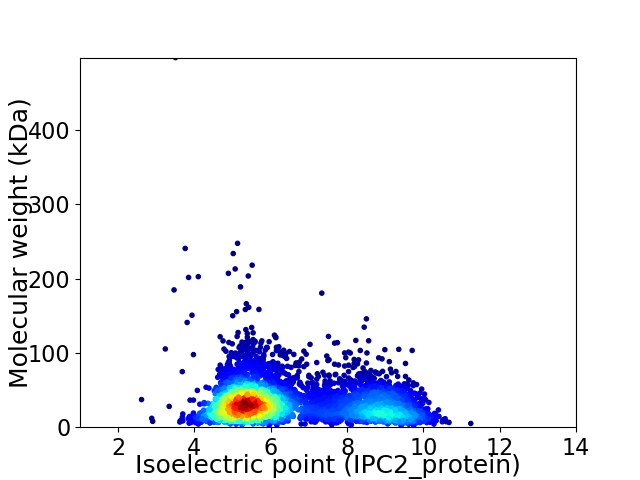
Virtual 2D-PAGE plot for 4688 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
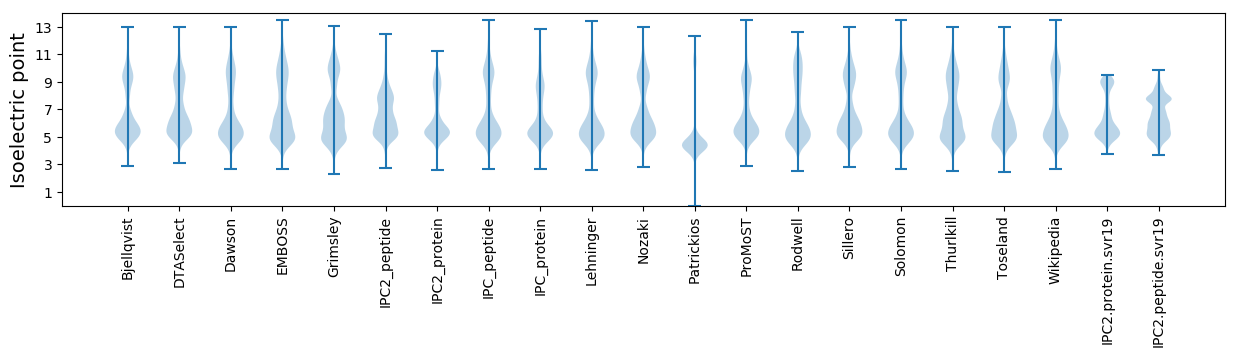
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q5RS48|A0A4Q5RS48_9SPHN Uncharacterized protein OS=Sphingomonadales bacterium OX=1978525 GN=EOP62_20475 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.24YY3 pKa = 10.02KK4 pKa = 10.81DD5 pKa = 3.42SGAVAGGEE13 pKa = 3.71ARR15 pKa = 11.84MITDD19 pKa = 3.24TLFRR23 pKa = 11.84VGGNDD28 pKa = 3.33EE29 pKa = 4.7GPGLTSDD36 pKa = 3.91VVRR39 pKa = 11.84TAGGPAYY46 pKa = 10.88YY47 pKa = 10.55NIAEE51 pKa = 4.44EE52 pKa = 4.43NGSPTSTVFGTLSHH66 pKa = 6.83SGLGYY71 pKa = 9.94QALGVDD77 pKa = 4.81FAGLSSTNVGPGAAFRR93 pKa = 11.84GIYY96 pKa = 8.03GTLHH100 pKa = 6.45LNSDD104 pKa = 3.65GSFGYY109 pKa = 11.1ALDD112 pKa = 3.97NTDD115 pKa = 4.56ADD117 pKa = 4.42TNQIAAGEE125 pKa = 4.34VTDD128 pKa = 4.32EE129 pKa = 4.32RR130 pKa = 11.84FTYY133 pKa = 9.7TYY135 pKa = 10.6QLNGQIFTDD144 pKa = 3.29KK145 pKa = 10.93VVFHH149 pKa = 6.3ITGVDD154 pKa = 3.2EE155 pKa = 4.96VGQQNHH161 pKa = 5.66YY162 pKa = 11.16LNTQMVRR169 pKa = 11.84TGDD172 pKa = 3.4MTIGANEE179 pKa = 4.01RR180 pKa = 11.84YY181 pKa = 9.93VFNTDD186 pKa = 3.12KK187 pKa = 11.3GISTASDD194 pKa = 3.08TTLDD198 pKa = 3.64VIHH201 pKa = 7.05NYY203 pKa = 10.26GSVRR207 pKa = 11.84VGGVSANGILAMTFDD222 pKa = 4.33PSQYY226 pKa = 10.75SEE228 pKa = 4.75GGSQLINDD236 pKa = 4.0GRR238 pKa = 11.84ITAISSLPSPANTGNNANAVSVSGITQPVTTNNGVIQAINTVTGAVFGAGALAIGAGNVINTGLVEE304 pKa = 4.63SYY306 pKa = 9.33STDD309 pKa = 3.08AGAGAINLGLYY320 pKa = 10.45SPTLINSGVIRR331 pKa = 11.84ASGGPRR337 pKa = 11.84ANPIEE342 pKa = 4.16ISAITAAGTGVHH354 pKa = 5.71VEE356 pKa = 3.85NSGTIEE362 pKa = 3.79AWEE365 pKa = 4.81DD366 pKa = 3.32NAGTQSIAFRR376 pKa = 11.84IFLNNTNVYY385 pKa = 9.62QGGIIANSGRR395 pKa = 11.84IVADD399 pKa = 3.18QAINALEE406 pKa = 4.72GIGGSLHH413 pKa = 6.51VINTGYY419 pKa = 10.09IQGDD423 pKa = 3.89YY424 pKa = 10.98VRR426 pKa = 11.84DD427 pKa = 3.96GVGHH431 pKa = 6.9DD432 pKa = 3.64VVTNAANGVWRR443 pKa = 11.84GNLGFATNADD453 pKa = 3.95LVRR456 pKa = 11.84NAGTVVGTISLGGGADD472 pKa = 3.21MFDD475 pKa = 3.48ARR477 pKa = 11.84TGGIVNGTVNGGAGNDD493 pKa = 3.52VLLGGIHH500 pKa = 7.59ADD502 pKa = 3.63TLIGGDD508 pKa = 3.58GADD511 pKa = 3.57ALFGGGGADD520 pKa = 3.55QLTGGAGADD529 pKa = 3.42LFIYY533 pKa = 10.33AVGDD537 pKa = 4.13SIVGARR543 pKa = 11.84DD544 pKa = 3.53TITDD548 pKa = 4.21FQTGQDD554 pKa = 3.85RR555 pKa = 11.84IDD557 pKa = 3.72LSRR560 pKa = 11.84LGVTSYY566 pKa = 11.55AITTEE571 pKa = 3.72GAYY574 pKa = 9.05TVLSGMGAGGAFNIAASGTIAASDD598 pKa = 4.8LIFAAQTSVMRR609 pKa = 11.84GGAGDD614 pKa = 4.54DD615 pKa = 3.69LLIAAMIGVTMDD627 pKa = 3.52GDD629 pKa = 3.58NGNDD633 pKa = 3.39VLVGSSGNDD642 pKa = 3.36VLDD645 pKa = 3.89GWNGADD651 pKa = 3.54TMFGGDD657 pKa = 3.5GDD659 pKa = 3.94DD660 pKa = 3.84TYY662 pKa = 12.01VVDD665 pKa = 3.85TDD667 pKa = 3.43GDD669 pKa = 3.84RR670 pKa = 11.84TIEE673 pKa = 4.12ADD675 pKa = 3.46GGGHH679 pKa = 7.12DD680 pKa = 4.83SVFSYY685 pKa = 10.77IDD687 pKa = 3.32YY688 pKa = 10.98SLQFWVEE695 pKa = 4.16DD696 pKa = 3.55GTLLGGSSSTIGLLGNRR713 pKa = 11.84LDD715 pKa = 3.54NTLRR719 pKa = 11.84GNAGSNRR726 pKa = 11.84ILGMDD731 pKa = 3.9GNDD734 pKa = 3.5VIIGGGGADD743 pKa = 3.89EE744 pKa = 4.82LFGGAGADD752 pKa = 3.14RR753 pKa = 11.84FVYY756 pKa = 10.15EE757 pKa = 4.28VASDD761 pKa = 3.51STAAAFDD768 pKa = 3.63WLKK771 pKa = 10.36MFEE774 pKa = 4.57HH775 pKa = 7.18GIDD778 pKa = 4.07SIDD781 pKa = 3.56LRR783 pKa = 11.84AIPVTTFNFEE793 pKa = 4.02PYY795 pKa = 9.65KK796 pKa = 10.89NSWQYY801 pKa = 9.82TEE803 pKa = 4.03WTTVTIGTTSGATMVMRR820 pKa = 11.84VDD822 pKa = 4.25GTATMADD829 pKa = 3.74FLVTARR835 pKa = 11.84GPTPNDD841 pKa = 4.54DD842 pKa = 3.96ILTGTNGVDD851 pKa = 3.99VIHH854 pKa = 6.74GLAGNDD860 pKa = 4.08TIDD863 pKa = 3.74GLLGADD869 pKa = 3.53QLYY872 pKa = 10.89GDD874 pKa = 4.82EE875 pKa = 4.85GNDD878 pKa = 3.25LFVFSAVQFGNTPDD892 pKa = 3.96LGLIDD897 pKa = 5.12GGADD901 pKa = 3.55YY902 pKa = 8.77DD903 pKa = 4.3TVDD906 pKa = 4.67LRR908 pKa = 11.84NISPAFVGTVALPGGGFSTALFVGSQRR935 pKa = 11.84YY936 pKa = 7.93QLTGVEE942 pKa = 5.17LILFGDD948 pKa = 4.73GEE950 pKa = 4.19NSLIGRR956 pKa = 11.84VSGSAPLEE964 pKa = 3.73IRR966 pKa = 11.84AGGGADD972 pKa = 4.23DD973 pKa = 4.31LTGSGGDD980 pKa = 3.28RR981 pKa = 11.84LYY983 pKa = 11.57GEE985 pKa = 5.22AGDD988 pKa = 4.4DD989 pKa = 3.4RR990 pKa = 11.84FFISGGFGVQPVSGVVDD1007 pKa = 4.28GGTGIDD1013 pKa = 3.44TLRR1016 pKa = 11.84LNNAFTVDD1024 pKa = 4.21LAAGTAVAGSYY1035 pKa = 9.09TYY1037 pKa = 10.13QVSGIEE1043 pKa = 4.2IVSAMTDD1050 pKa = 3.22SVVRR1054 pKa = 11.84GDD1056 pKa = 3.37AGANIFNVNPLFDD1069 pKa = 3.78STSASVFFDD1078 pKa = 2.99GRR1080 pKa = 11.84EE1081 pKa = 4.03GDD1083 pKa = 3.99DD1084 pKa = 3.73SLTGSMGNDD1093 pKa = 3.24QLIGGAGNDD1102 pKa = 3.92TLVGGAGADD1111 pKa = 3.35AMTGGSGNDD1120 pKa = 3.71LYY1122 pKa = 11.65SVDD1125 pKa = 3.61NAGDD1129 pKa = 3.67VVTEE1133 pKa = 4.07LLGEE1137 pKa = 4.68GYY1139 pKa = 10.7DD1140 pKa = 3.68EE1141 pKa = 4.56VSASISYY1148 pKa = 10.6VLTANVEE1155 pKa = 4.07GLRR1158 pKa = 11.84LVNGSGALNGTGNALDD1174 pKa = 4.1NLITGNDD1181 pKa = 3.59GANRR1185 pKa = 11.84LDD1187 pKa = 4.19GGDD1190 pKa = 3.88GNDD1193 pKa = 4.23ILWSSDD1199 pKa = 2.93GTSDD1203 pKa = 5.23DD1204 pKa = 3.64VGLEE1208 pKa = 3.75HH1209 pKa = 7.52DD1210 pKa = 4.29QLFGGAGNDD1219 pKa = 3.39ALAVGVGDD1227 pKa = 4.87DD1228 pKa = 3.93ADD1230 pKa = 4.17GGSGVDD1236 pKa = 3.46TLRR1239 pKa = 11.84LSLAGFSWGVTLDD1252 pKa = 3.39MSGFTSGGAFNIFGGIISEE1271 pKa = 4.38IEE1273 pKa = 4.04VIDD1276 pKa = 4.43YY1277 pKa = 10.77IVGSAGNDD1285 pKa = 3.29KK1286 pKa = 9.37ITLGGHH1292 pKa = 5.53TYY1294 pKa = 10.91GVTVKK1299 pKa = 10.42SGAGNDD1305 pKa = 3.86TLLSGAGADD1314 pKa = 3.69HH1315 pKa = 7.4LYY1317 pKa = 11.07GGLGDD1322 pKa = 4.67DD1323 pKa = 4.1VFVVDD1328 pKa = 4.59TGDD1331 pKa = 3.92DD1332 pKa = 3.79LVFEE1336 pKa = 4.44NAGEE1340 pKa = 4.27GFDD1343 pKa = 3.3IVYY1346 pKa = 10.45AAGSYY1351 pKa = 10.83ALVAGSHH1358 pKa = 5.65VEE1360 pKa = 3.92LLTTLSVDD1368 pKa = 3.61GAVTLVGNEE1377 pKa = 3.85QANTILGNAGANYY1390 pKa = 10.11LAGGAGADD1398 pKa = 3.51TLAGNGGNDD1407 pKa = 3.26VMFGEE1412 pKa = 5.75DD1413 pKa = 3.97GDD1415 pKa = 5.24DD1416 pKa = 5.76LIGDD1420 pKa = 4.27DD1421 pKa = 5.43AGDD1424 pKa = 4.27DD1425 pKa = 3.95YY1426 pKa = 12.22AEE1428 pKa = 4.66GGAGNDD1434 pKa = 3.75YY1435 pKa = 10.8IYY1437 pKa = 10.88LGAGNDD1443 pKa = 3.52RR1444 pKa = 11.84LYY1446 pKa = 11.41GGAGNDD1452 pKa = 3.95LLFGQDD1458 pKa = 3.65GDD1460 pKa = 4.19DD1461 pKa = 3.69TLGDD1465 pKa = 3.72GDD1467 pKa = 4.98GDD1469 pKa = 4.11DD1470 pKa = 4.37MLDD1473 pKa = 3.53GGEE1476 pKa = 4.32GNDD1479 pKa = 3.59YY1480 pKa = 10.88LYY1482 pKa = 11.16AGIGNDD1488 pKa = 3.11RR1489 pKa = 11.84LYY1491 pKa = 11.36GGGGDD1496 pKa = 3.91DD1497 pKa = 5.18QMFGQDD1503 pKa = 3.88GDD1505 pKa = 4.78DD1506 pKa = 3.34IMGGGGGADD1515 pKa = 3.53YY1516 pKa = 10.72MEE1518 pKa = 5.4AGAGKK1523 pKa = 10.04DD1524 pKa = 3.34FAYY1527 pKa = 10.4GGDD1530 pKa = 3.81GNDD1533 pKa = 3.22VVLGEE1538 pKa = 4.85AGDD1541 pKa = 3.81DD1542 pKa = 3.52QLYY1545 pKa = 11.03GEE1547 pKa = 5.31GGDD1550 pKa = 4.91DD1551 pKa = 3.52ILIGGTGNDD1560 pKa = 3.3QLFGGEE1566 pKa = 4.22GADD1569 pKa = 3.34ILGDD1573 pKa = 4.01AEE1575 pKa = 5.1GDD1577 pKa = 3.75DD1578 pKa = 4.08YY1579 pKa = 11.5MQGGGGADD1587 pKa = 3.52YY1588 pKa = 10.66LYY1590 pKa = 10.93AGAGRR1595 pKa = 11.84DD1596 pKa = 3.51TLYY1599 pKa = 11.38GDD1601 pKa = 4.43GGNDD1605 pKa = 2.86ILFGQDD1611 pKa = 3.3GDD1613 pKa = 4.84DD1614 pKa = 5.06LLGDD1618 pKa = 4.22DD1619 pKa = 5.11EE1620 pKa = 7.2GDD1622 pKa = 3.78DD1623 pKa = 4.38VLDD1626 pKa = 4.28GGNGNDD1632 pKa = 3.79YY1633 pKa = 10.11MFAGAGADD1641 pKa = 3.44RR1642 pKa = 11.84LYY1644 pKa = 11.38GGAGDD1649 pKa = 5.57DD1650 pKa = 5.28LMFGQDD1656 pKa = 4.55GDD1658 pKa = 4.64DD1659 pKa = 3.77EE1660 pKa = 4.48MAGGDD1665 pKa = 4.13GADD1668 pKa = 4.01MLDD1671 pKa = 3.68GGAGADD1677 pKa = 3.6YY1678 pKa = 10.44LYY1680 pKa = 11.02SGTGNDD1686 pKa = 3.45RR1687 pKa = 11.84LYY1689 pKa = 11.34GGDD1692 pKa = 4.06GNDD1695 pKa = 4.14LMFGQEE1701 pKa = 4.53GDD1703 pKa = 4.06DD1704 pKa = 3.94EE1705 pKa = 4.71LAGGAGADD1713 pKa = 3.98YY1714 pKa = 11.08IEE1716 pKa = 5.44GGDD1719 pKa = 4.26GNDD1722 pKa = 4.7LIYY1725 pKa = 11.06GGAGTDD1731 pKa = 3.42MLYY1734 pKa = 11.08GGAGADD1740 pKa = 3.21AFAFMAAPGAANLQTIGDD1758 pKa = 4.0FTVGQDD1764 pKa = 3.15RR1765 pKa = 11.84LLLSGAVFAGLAPGALAAGAFVTGTAAADD1794 pKa = 3.28ASDD1797 pKa = 4.48RR1798 pKa = 11.84ILYY1801 pKa = 9.22NAATGALWFDD1811 pKa = 3.79ADD1813 pKa = 3.76GTGAIAAVQFGMLSPGLALSATDD1836 pKa = 3.6FAVII1840 pKa = 4.32
MM1 pKa = 7.73KK2 pKa = 10.24YY3 pKa = 10.02KK4 pKa = 10.81DD5 pKa = 3.42SGAVAGGEE13 pKa = 3.71ARR15 pKa = 11.84MITDD19 pKa = 3.24TLFRR23 pKa = 11.84VGGNDD28 pKa = 3.33EE29 pKa = 4.7GPGLTSDD36 pKa = 3.91VVRR39 pKa = 11.84TAGGPAYY46 pKa = 10.88YY47 pKa = 10.55NIAEE51 pKa = 4.44EE52 pKa = 4.43NGSPTSTVFGTLSHH66 pKa = 6.83SGLGYY71 pKa = 9.94QALGVDD77 pKa = 4.81FAGLSSTNVGPGAAFRR93 pKa = 11.84GIYY96 pKa = 8.03GTLHH100 pKa = 6.45LNSDD104 pKa = 3.65GSFGYY109 pKa = 11.1ALDD112 pKa = 3.97NTDD115 pKa = 4.56ADD117 pKa = 4.42TNQIAAGEE125 pKa = 4.34VTDD128 pKa = 4.32EE129 pKa = 4.32RR130 pKa = 11.84FTYY133 pKa = 9.7TYY135 pKa = 10.6QLNGQIFTDD144 pKa = 3.29KK145 pKa = 10.93VVFHH149 pKa = 6.3ITGVDD154 pKa = 3.2EE155 pKa = 4.96VGQQNHH161 pKa = 5.66YY162 pKa = 11.16LNTQMVRR169 pKa = 11.84TGDD172 pKa = 3.4MTIGANEE179 pKa = 4.01RR180 pKa = 11.84YY181 pKa = 9.93VFNTDD186 pKa = 3.12KK187 pKa = 11.3GISTASDD194 pKa = 3.08TTLDD198 pKa = 3.64VIHH201 pKa = 7.05NYY203 pKa = 10.26GSVRR207 pKa = 11.84VGGVSANGILAMTFDD222 pKa = 4.33PSQYY226 pKa = 10.75SEE228 pKa = 4.75GGSQLINDD236 pKa = 4.0GRR238 pKa = 11.84ITAISSLPSPANTGNNANAVSVSGITQPVTTNNGVIQAINTVTGAVFGAGALAIGAGNVINTGLVEE304 pKa = 4.63SYY306 pKa = 9.33STDD309 pKa = 3.08AGAGAINLGLYY320 pKa = 10.45SPTLINSGVIRR331 pKa = 11.84ASGGPRR337 pKa = 11.84ANPIEE342 pKa = 4.16ISAITAAGTGVHH354 pKa = 5.71VEE356 pKa = 3.85NSGTIEE362 pKa = 3.79AWEE365 pKa = 4.81DD366 pKa = 3.32NAGTQSIAFRR376 pKa = 11.84IFLNNTNVYY385 pKa = 9.62QGGIIANSGRR395 pKa = 11.84IVADD399 pKa = 3.18QAINALEE406 pKa = 4.72GIGGSLHH413 pKa = 6.51VINTGYY419 pKa = 10.09IQGDD423 pKa = 3.89YY424 pKa = 10.98VRR426 pKa = 11.84DD427 pKa = 3.96GVGHH431 pKa = 6.9DD432 pKa = 3.64VVTNAANGVWRR443 pKa = 11.84GNLGFATNADD453 pKa = 3.95LVRR456 pKa = 11.84NAGTVVGTISLGGGADD472 pKa = 3.21MFDD475 pKa = 3.48ARR477 pKa = 11.84TGGIVNGTVNGGAGNDD493 pKa = 3.52VLLGGIHH500 pKa = 7.59ADD502 pKa = 3.63TLIGGDD508 pKa = 3.58GADD511 pKa = 3.57ALFGGGGADD520 pKa = 3.55QLTGGAGADD529 pKa = 3.42LFIYY533 pKa = 10.33AVGDD537 pKa = 4.13SIVGARR543 pKa = 11.84DD544 pKa = 3.53TITDD548 pKa = 4.21FQTGQDD554 pKa = 3.85RR555 pKa = 11.84IDD557 pKa = 3.72LSRR560 pKa = 11.84LGVTSYY566 pKa = 11.55AITTEE571 pKa = 3.72GAYY574 pKa = 9.05TVLSGMGAGGAFNIAASGTIAASDD598 pKa = 4.8LIFAAQTSVMRR609 pKa = 11.84GGAGDD614 pKa = 4.54DD615 pKa = 3.69LLIAAMIGVTMDD627 pKa = 3.52GDD629 pKa = 3.58NGNDD633 pKa = 3.39VLVGSSGNDD642 pKa = 3.36VLDD645 pKa = 3.89GWNGADD651 pKa = 3.54TMFGGDD657 pKa = 3.5GDD659 pKa = 3.94DD660 pKa = 3.84TYY662 pKa = 12.01VVDD665 pKa = 3.85TDD667 pKa = 3.43GDD669 pKa = 3.84RR670 pKa = 11.84TIEE673 pKa = 4.12ADD675 pKa = 3.46GGGHH679 pKa = 7.12DD680 pKa = 4.83SVFSYY685 pKa = 10.77IDD687 pKa = 3.32YY688 pKa = 10.98SLQFWVEE695 pKa = 4.16DD696 pKa = 3.55GTLLGGSSSTIGLLGNRR713 pKa = 11.84LDD715 pKa = 3.54NTLRR719 pKa = 11.84GNAGSNRR726 pKa = 11.84ILGMDD731 pKa = 3.9GNDD734 pKa = 3.5VIIGGGGADD743 pKa = 3.89EE744 pKa = 4.82LFGGAGADD752 pKa = 3.14RR753 pKa = 11.84FVYY756 pKa = 10.15EE757 pKa = 4.28VASDD761 pKa = 3.51STAAAFDD768 pKa = 3.63WLKK771 pKa = 10.36MFEE774 pKa = 4.57HH775 pKa = 7.18GIDD778 pKa = 4.07SIDD781 pKa = 3.56LRR783 pKa = 11.84AIPVTTFNFEE793 pKa = 4.02PYY795 pKa = 9.65KK796 pKa = 10.89NSWQYY801 pKa = 9.82TEE803 pKa = 4.03WTTVTIGTTSGATMVMRR820 pKa = 11.84VDD822 pKa = 4.25GTATMADD829 pKa = 3.74FLVTARR835 pKa = 11.84GPTPNDD841 pKa = 4.54DD842 pKa = 3.96ILTGTNGVDD851 pKa = 3.99VIHH854 pKa = 6.74GLAGNDD860 pKa = 4.08TIDD863 pKa = 3.74GLLGADD869 pKa = 3.53QLYY872 pKa = 10.89GDD874 pKa = 4.82EE875 pKa = 4.85GNDD878 pKa = 3.25LFVFSAVQFGNTPDD892 pKa = 3.96LGLIDD897 pKa = 5.12GGADD901 pKa = 3.55YY902 pKa = 8.77DD903 pKa = 4.3TVDD906 pKa = 4.67LRR908 pKa = 11.84NISPAFVGTVALPGGGFSTALFVGSQRR935 pKa = 11.84YY936 pKa = 7.93QLTGVEE942 pKa = 5.17LILFGDD948 pKa = 4.73GEE950 pKa = 4.19NSLIGRR956 pKa = 11.84VSGSAPLEE964 pKa = 3.73IRR966 pKa = 11.84AGGGADD972 pKa = 4.23DD973 pKa = 4.31LTGSGGDD980 pKa = 3.28RR981 pKa = 11.84LYY983 pKa = 11.57GEE985 pKa = 5.22AGDD988 pKa = 4.4DD989 pKa = 3.4RR990 pKa = 11.84FFISGGFGVQPVSGVVDD1007 pKa = 4.28GGTGIDD1013 pKa = 3.44TLRR1016 pKa = 11.84LNNAFTVDD1024 pKa = 4.21LAAGTAVAGSYY1035 pKa = 9.09TYY1037 pKa = 10.13QVSGIEE1043 pKa = 4.2IVSAMTDD1050 pKa = 3.22SVVRR1054 pKa = 11.84GDD1056 pKa = 3.37AGANIFNVNPLFDD1069 pKa = 3.78STSASVFFDD1078 pKa = 2.99GRR1080 pKa = 11.84EE1081 pKa = 4.03GDD1083 pKa = 3.99DD1084 pKa = 3.73SLTGSMGNDD1093 pKa = 3.24QLIGGAGNDD1102 pKa = 3.92TLVGGAGADD1111 pKa = 3.35AMTGGSGNDD1120 pKa = 3.71LYY1122 pKa = 11.65SVDD1125 pKa = 3.61NAGDD1129 pKa = 3.67VVTEE1133 pKa = 4.07LLGEE1137 pKa = 4.68GYY1139 pKa = 10.7DD1140 pKa = 3.68EE1141 pKa = 4.56VSASISYY1148 pKa = 10.6VLTANVEE1155 pKa = 4.07GLRR1158 pKa = 11.84LVNGSGALNGTGNALDD1174 pKa = 4.1NLITGNDD1181 pKa = 3.59GANRR1185 pKa = 11.84LDD1187 pKa = 4.19GGDD1190 pKa = 3.88GNDD1193 pKa = 4.23ILWSSDD1199 pKa = 2.93GTSDD1203 pKa = 5.23DD1204 pKa = 3.64VGLEE1208 pKa = 3.75HH1209 pKa = 7.52DD1210 pKa = 4.29QLFGGAGNDD1219 pKa = 3.39ALAVGVGDD1227 pKa = 4.87DD1228 pKa = 3.93ADD1230 pKa = 4.17GGSGVDD1236 pKa = 3.46TLRR1239 pKa = 11.84LSLAGFSWGVTLDD1252 pKa = 3.39MSGFTSGGAFNIFGGIISEE1271 pKa = 4.38IEE1273 pKa = 4.04VIDD1276 pKa = 4.43YY1277 pKa = 10.77IVGSAGNDD1285 pKa = 3.29KK1286 pKa = 9.37ITLGGHH1292 pKa = 5.53TYY1294 pKa = 10.91GVTVKK1299 pKa = 10.42SGAGNDD1305 pKa = 3.86TLLSGAGADD1314 pKa = 3.69HH1315 pKa = 7.4LYY1317 pKa = 11.07GGLGDD1322 pKa = 4.67DD1323 pKa = 4.1VFVVDD1328 pKa = 4.59TGDD1331 pKa = 3.92DD1332 pKa = 3.79LVFEE1336 pKa = 4.44NAGEE1340 pKa = 4.27GFDD1343 pKa = 3.3IVYY1346 pKa = 10.45AAGSYY1351 pKa = 10.83ALVAGSHH1358 pKa = 5.65VEE1360 pKa = 3.92LLTTLSVDD1368 pKa = 3.61GAVTLVGNEE1377 pKa = 3.85QANTILGNAGANYY1390 pKa = 10.11LAGGAGADD1398 pKa = 3.51TLAGNGGNDD1407 pKa = 3.26VMFGEE1412 pKa = 5.75DD1413 pKa = 3.97GDD1415 pKa = 5.24DD1416 pKa = 5.76LIGDD1420 pKa = 4.27DD1421 pKa = 5.43AGDD1424 pKa = 4.27DD1425 pKa = 3.95YY1426 pKa = 12.22AEE1428 pKa = 4.66GGAGNDD1434 pKa = 3.75YY1435 pKa = 10.8IYY1437 pKa = 10.88LGAGNDD1443 pKa = 3.52RR1444 pKa = 11.84LYY1446 pKa = 11.41GGAGNDD1452 pKa = 3.95LLFGQDD1458 pKa = 3.65GDD1460 pKa = 4.19DD1461 pKa = 3.69TLGDD1465 pKa = 3.72GDD1467 pKa = 4.98GDD1469 pKa = 4.11DD1470 pKa = 4.37MLDD1473 pKa = 3.53GGEE1476 pKa = 4.32GNDD1479 pKa = 3.59YY1480 pKa = 10.88LYY1482 pKa = 11.16AGIGNDD1488 pKa = 3.11RR1489 pKa = 11.84LYY1491 pKa = 11.36GGGGDD1496 pKa = 3.91DD1497 pKa = 5.18QMFGQDD1503 pKa = 3.88GDD1505 pKa = 4.78DD1506 pKa = 3.34IMGGGGGADD1515 pKa = 3.53YY1516 pKa = 10.72MEE1518 pKa = 5.4AGAGKK1523 pKa = 10.04DD1524 pKa = 3.34FAYY1527 pKa = 10.4GGDD1530 pKa = 3.81GNDD1533 pKa = 3.22VVLGEE1538 pKa = 4.85AGDD1541 pKa = 3.81DD1542 pKa = 3.52QLYY1545 pKa = 11.03GEE1547 pKa = 5.31GGDD1550 pKa = 4.91DD1551 pKa = 3.52ILIGGTGNDD1560 pKa = 3.3QLFGGEE1566 pKa = 4.22GADD1569 pKa = 3.34ILGDD1573 pKa = 4.01AEE1575 pKa = 5.1GDD1577 pKa = 3.75DD1578 pKa = 4.08YY1579 pKa = 11.5MQGGGGADD1587 pKa = 3.52YY1588 pKa = 10.66LYY1590 pKa = 10.93AGAGRR1595 pKa = 11.84DD1596 pKa = 3.51TLYY1599 pKa = 11.38GDD1601 pKa = 4.43GGNDD1605 pKa = 2.86ILFGQDD1611 pKa = 3.3GDD1613 pKa = 4.84DD1614 pKa = 5.06LLGDD1618 pKa = 4.22DD1619 pKa = 5.11EE1620 pKa = 7.2GDD1622 pKa = 3.78DD1623 pKa = 4.38VLDD1626 pKa = 4.28GGNGNDD1632 pKa = 3.79YY1633 pKa = 10.11MFAGAGADD1641 pKa = 3.44RR1642 pKa = 11.84LYY1644 pKa = 11.38GGAGDD1649 pKa = 5.57DD1650 pKa = 5.28LMFGQDD1656 pKa = 4.55GDD1658 pKa = 4.64DD1659 pKa = 3.77EE1660 pKa = 4.48MAGGDD1665 pKa = 4.13GADD1668 pKa = 4.01MLDD1671 pKa = 3.68GGAGADD1677 pKa = 3.6YY1678 pKa = 10.44LYY1680 pKa = 11.02SGTGNDD1686 pKa = 3.45RR1687 pKa = 11.84LYY1689 pKa = 11.34GGDD1692 pKa = 4.06GNDD1695 pKa = 4.14LMFGQEE1701 pKa = 4.53GDD1703 pKa = 4.06DD1704 pKa = 3.94EE1705 pKa = 4.71LAGGAGADD1713 pKa = 3.98YY1714 pKa = 11.08IEE1716 pKa = 5.44GGDD1719 pKa = 4.26GNDD1722 pKa = 4.7LIYY1725 pKa = 11.06GGAGTDD1731 pKa = 3.42MLYY1734 pKa = 11.08GGAGADD1740 pKa = 3.21AFAFMAAPGAANLQTIGDD1758 pKa = 4.0FTVGQDD1764 pKa = 3.15RR1765 pKa = 11.84LLLSGAVFAGLAPGALAAGAFVTGTAAADD1794 pKa = 3.28ASDD1797 pKa = 4.48RR1798 pKa = 11.84ILYY1801 pKa = 9.22NAATGALWFDD1811 pKa = 3.79ADD1813 pKa = 3.76GTGAIAAVQFGMLSPGLALSATDD1836 pKa = 3.6FAVII1840 pKa = 4.32
Molecular weight: 184.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q5RYX5|A0A4Q5RYX5_9SPHN Uncharacterized protein OS=Sphingomonadales bacterium OX=1978525 GN=EOP62_08285 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.09VIHH32 pKa = 6.46ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84SKK41 pKa = 11.12LSAA44 pKa = 3.81
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.09VIHH32 pKa = 6.46ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84SKK41 pKa = 11.12LSAA44 pKa = 3.81
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1519867 |
22 |
4969 |
324.2 |
35.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.074 ± 0.047 | 0.757 ± 0.011 |
5.838 ± 0.033 | 5.454 ± 0.038 |
3.661 ± 0.022 | 8.965 ± 0.045 |
1.939 ± 0.019 | 5.181 ± 0.025 |
3.159 ± 0.027 | 9.686 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.418 ± 0.02 | 2.722 ± 0.031 |
5.227 ± 0.033 | 3.042 ± 0.02 |
7.046 ± 0.044 | 5.474 ± 0.027 |
5.48 ± 0.052 | 7.131 ± 0.029 |
1.459 ± 0.018 | 2.289 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
