
Effusibacillus lacus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Alicyclobacillaceae; Effusibacillus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
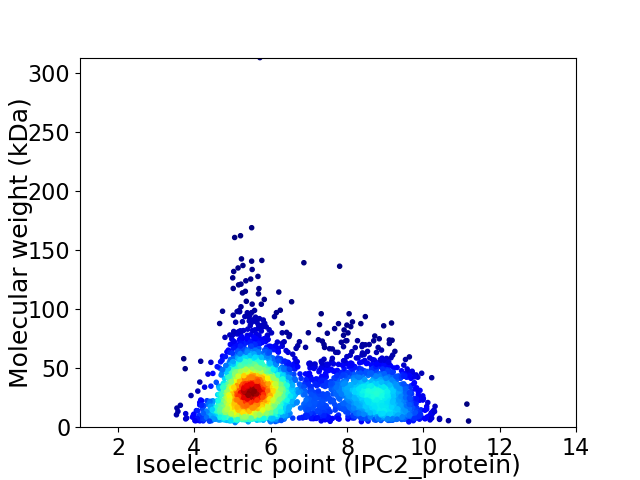
Virtual 2D-PAGE plot for 3692 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A292YLW0|A0A292YLW0_9BACL Glutamate synthase subunit alpha OS=Effusibacillus lacus OX=1348429 GN=EFBL_2570 PE=3 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84FMRR5 pKa = 11.84TIGCTLALVSLLAAPPVADD24 pKa = 4.05AQSLSIVSITVNGKK38 pKa = 7.7QVPLSDD44 pKa = 3.69SVPYY48 pKa = 10.49LDD50 pKa = 4.57SEE52 pKa = 4.74SGRR55 pKa = 11.84VMVPIRR61 pKa = 11.84FVAEE65 pKa = 3.78HH66 pKa = 6.2MGAAVNWNEE75 pKa = 4.08SAQQATIVLNSRR87 pKa = 11.84QITLTIGQKK96 pKa = 7.2TAYY99 pKa = 10.3VNWQPYY105 pKa = 7.32QLEE108 pKa = 4.29TAPVLRR114 pKa = 11.84NGRR117 pKa = 11.84TYY119 pKa = 10.77VPVRR123 pKa = 11.84FVGEE127 pKa = 3.74ALGANLYY134 pKa = 8.1WHH136 pKa = 7.36EE137 pKa = 4.09NTKK140 pKa = 9.13TVEE143 pKa = 3.8IVQNNGSSTPDD154 pKa = 3.1YY155 pKa = 10.73SDD157 pKa = 4.51PYY159 pKa = 10.58TPGTPSGDD167 pKa = 3.31QSDD170 pKa = 5.0YY171 pKa = 11.68GDD173 pKa = 3.85DD174 pKa = 3.42WDD176 pKa = 4.42TFSHH180 pKa = 6.52KK181 pKa = 9.54WDD183 pKa = 4.2EE184 pKa = 4.11YY185 pKa = 10.07SIQYY189 pKa = 9.48PEE191 pKa = 5.12DD192 pKa = 3.35WPKK195 pKa = 10.29PRR197 pKa = 11.84VVPGYY202 pKa = 10.0VKK204 pKa = 10.45WEE206 pKa = 4.21LDD208 pKa = 4.03DD209 pKa = 5.85DD210 pKa = 5.82DD211 pKa = 5.13EE212 pKa = 5.53DD213 pKa = 5.04AYY215 pKa = 10.98AYY217 pKa = 9.82IRR219 pKa = 11.84FSRR222 pKa = 11.84TTPVSNPIGEE232 pKa = 4.97DD233 pKa = 3.28DD234 pKa = 5.44ADD236 pKa = 5.17PIDD239 pKa = 5.57LNDD242 pKa = 3.95GTDD245 pKa = 3.33AEE247 pKa = 4.57YY248 pKa = 10.88EE249 pKa = 3.77KK250 pKa = 11.16DD251 pKa = 3.4EE252 pKa = 4.17YY253 pKa = 11.6SNRR256 pKa = 11.84TVYY259 pKa = 9.86MMRR262 pKa = 11.84VDD264 pKa = 3.51EE265 pKa = 4.76GDD267 pKa = 3.83YY268 pKa = 11.09EE269 pKa = 4.17ATLYY273 pKa = 11.1VSLPDD278 pKa = 5.98DD279 pKa = 3.97MDD281 pKa = 5.91DD282 pKa = 5.45DD283 pKa = 4.78YY284 pKa = 11.75DD285 pKa = 3.6IKK287 pKa = 11.54KK288 pKa = 8.78MFRR291 pKa = 11.84SFEE294 pKa = 4.08VEE296 pKa = 3.98EE297 pKa = 4.12EE298 pKa = 4.29DD299 pKa = 4.76GNEE302 pKa = 4.18DD303 pKa = 3.84LVDD306 pKa = 3.89YY307 pKa = 11.52VNTKK311 pKa = 10.47YY312 pKa = 10.47FFSLQYY318 pKa = 10.18PEE320 pKa = 4.74EE321 pKa = 4.3WDD323 pKa = 5.11DD324 pKa = 3.97GDD326 pKa = 4.72NDD328 pKa = 4.15VDD330 pKa = 3.71THH332 pKa = 7.34SNGIEE337 pKa = 3.98WDD339 pKa = 3.57LSHH342 pKa = 7.57ADD344 pKa = 3.62VEE346 pKa = 5.21FYY348 pKa = 10.68AYY350 pKa = 10.25DD351 pKa = 4.2EE352 pKa = 5.16EE353 pKa = 5.99DD354 pKa = 4.7LDD356 pKa = 4.46WDD358 pKa = 4.92DD359 pKa = 6.65LEE361 pKa = 6.94DD362 pKa = 5.14DD363 pKa = 4.18YY364 pKa = 12.24ADD366 pKa = 3.88ADD368 pKa = 3.97EE369 pKa = 4.83EE370 pKa = 4.2EE371 pKa = 5.03LRR373 pKa = 11.84DD374 pKa = 3.86GTDD377 pKa = 2.79AYY379 pKa = 11.45LWIDD383 pKa = 3.7DD384 pKa = 4.31DD385 pKa = 4.72GSYY388 pKa = 11.13VDD390 pKa = 3.81YY391 pKa = 11.62VFFFRR396 pKa = 11.84KK397 pKa = 9.8NDD399 pKa = 3.31VVYY402 pKa = 10.5KK403 pKa = 10.7GEE405 pKa = 4.02ARR407 pKa = 11.84VHH409 pKa = 6.46EE410 pKa = 5.1DD411 pKa = 3.34YY412 pKa = 10.92QDD414 pKa = 3.53EE415 pKa = 4.59YY416 pKa = 11.16EE417 pKa = 5.8DD418 pKa = 5.1EE419 pKa = 4.2ILDD422 pKa = 3.67MFKK425 pKa = 11.01SVVADD430 pKa = 3.68
MM1 pKa = 7.81RR2 pKa = 11.84FMRR5 pKa = 11.84TIGCTLALVSLLAAPPVADD24 pKa = 4.05AQSLSIVSITVNGKK38 pKa = 7.7QVPLSDD44 pKa = 3.69SVPYY48 pKa = 10.49LDD50 pKa = 4.57SEE52 pKa = 4.74SGRR55 pKa = 11.84VMVPIRR61 pKa = 11.84FVAEE65 pKa = 3.78HH66 pKa = 6.2MGAAVNWNEE75 pKa = 4.08SAQQATIVLNSRR87 pKa = 11.84QITLTIGQKK96 pKa = 7.2TAYY99 pKa = 10.3VNWQPYY105 pKa = 7.32QLEE108 pKa = 4.29TAPVLRR114 pKa = 11.84NGRR117 pKa = 11.84TYY119 pKa = 10.77VPVRR123 pKa = 11.84FVGEE127 pKa = 3.74ALGANLYY134 pKa = 8.1WHH136 pKa = 7.36EE137 pKa = 4.09NTKK140 pKa = 9.13TVEE143 pKa = 3.8IVQNNGSSTPDD154 pKa = 3.1YY155 pKa = 10.73SDD157 pKa = 4.51PYY159 pKa = 10.58TPGTPSGDD167 pKa = 3.31QSDD170 pKa = 5.0YY171 pKa = 11.68GDD173 pKa = 3.85DD174 pKa = 3.42WDD176 pKa = 4.42TFSHH180 pKa = 6.52KK181 pKa = 9.54WDD183 pKa = 4.2EE184 pKa = 4.11YY185 pKa = 10.07SIQYY189 pKa = 9.48PEE191 pKa = 5.12DD192 pKa = 3.35WPKK195 pKa = 10.29PRR197 pKa = 11.84VVPGYY202 pKa = 10.0VKK204 pKa = 10.45WEE206 pKa = 4.21LDD208 pKa = 4.03DD209 pKa = 5.85DD210 pKa = 5.82DD211 pKa = 5.13EE212 pKa = 5.53DD213 pKa = 5.04AYY215 pKa = 10.98AYY217 pKa = 9.82IRR219 pKa = 11.84FSRR222 pKa = 11.84TTPVSNPIGEE232 pKa = 4.97DD233 pKa = 3.28DD234 pKa = 5.44ADD236 pKa = 5.17PIDD239 pKa = 5.57LNDD242 pKa = 3.95GTDD245 pKa = 3.33AEE247 pKa = 4.57YY248 pKa = 10.88EE249 pKa = 3.77KK250 pKa = 11.16DD251 pKa = 3.4EE252 pKa = 4.17YY253 pKa = 11.6SNRR256 pKa = 11.84TVYY259 pKa = 9.86MMRR262 pKa = 11.84VDD264 pKa = 3.51EE265 pKa = 4.76GDD267 pKa = 3.83YY268 pKa = 11.09EE269 pKa = 4.17ATLYY273 pKa = 11.1VSLPDD278 pKa = 5.98DD279 pKa = 3.97MDD281 pKa = 5.91DD282 pKa = 5.45DD283 pKa = 4.78YY284 pKa = 11.75DD285 pKa = 3.6IKK287 pKa = 11.54KK288 pKa = 8.78MFRR291 pKa = 11.84SFEE294 pKa = 4.08VEE296 pKa = 3.98EE297 pKa = 4.12EE298 pKa = 4.29DD299 pKa = 4.76GNEE302 pKa = 4.18DD303 pKa = 3.84LVDD306 pKa = 3.89YY307 pKa = 11.52VNTKK311 pKa = 10.47YY312 pKa = 10.47FFSLQYY318 pKa = 10.18PEE320 pKa = 4.74EE321 pKa = 4.3WDD323 pKa = 5.11DD324 pKa = 3.97GDD326 pKa = 4.72NDD328 pKa = 4.15VDD330 pKa = 3.71THH332 pKa = 7.34SNGIEE337 pKa = 3.98WDD339 pKa = 3.57LSHH342 pKa = 7.57ADD344 pKa = 3.62VEE346 pKa = 5.21FYY348 pKa = 10.68AYY350 pKa = 10.25DD351 pKa = 4.2EE352 pKa = 5.16EE353 pKa = 5.99DD354 pKa = 4.7LDD356 pKa = 4.46WDD358 pKa = 4.92DD359 pKa = 6.65LEE361 pKa = 6.94DD362 pKa = 5.14DD363 pKa = 4.18YY364 pKa = 12.24ADD366 pKa = 3.88ADD368 pKa = 3.97EE369 pKa = 4.83EE370 pKa = 4.2EE371 pKa = 5.03LRR373 pKa = 11.84DD374 pKa = 3.86GTDD377 pKa = 2.79AYY379 pKa = 11.45LWIDD383 pKa = 3.7DD384 pKa = 4.31DD385 pKa = 4.72GSYY388 pKa = 11.13VDD390 pKa = 3.81YY391 pKa = 11.62VFFFRR396 pKa = 11.84KK397 pKa = 9.8NDD399 pKa = 3.31VVYY402 pKa = 10.5KK403 pKa = 10.7GEE405 pKa = 4.02ARR407 pKa = 11.84VHH409 pKa = 6.46EE410 pKa = 5.1DD411 pKa = 3.34YY412 pKa = 10.92QDD414 pKa = 3.53EE415 pKa = 4.59YY416 pKa = 11.16EE417 pKa = 5.8DD418 pKa = 5.1EE419 pKa = 4.2ILDD422 pKa = 3.67MFKK425 pKa = 11.01SVVADD430 pKa = 3.68
Molecular weight: 49.49 kDa
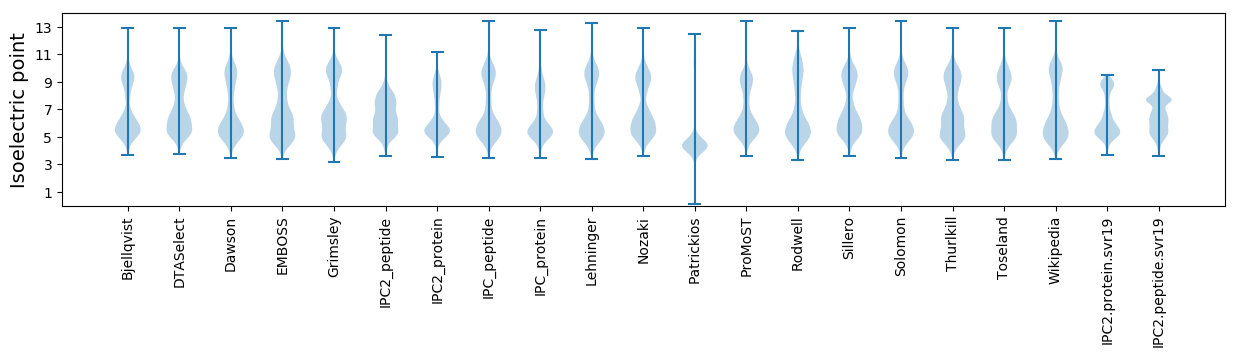
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A292YCK6|A0A292YCK6_9BACL Branched-chain amino acid ABC transporter permease OS=Effusibacillus lacus OX=1348429 GN=EFBL_0880 PE=4 SV=1
MM1 pKa = 7.6KK2 pKa = 8.69PTFQPNVRR10 pKa = 11.84KK11 pKa = 9.76RR12 pKa = 11.84KK13 pKa = 8.88KK14 pKa = 8.64VHH16 pKa = 5.57GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.91VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
MM1 pKa = 7.6KK2 pKa = 8.69PTFQPNVRR10 pKa = 11.84KK11 pKa = 9.76RR12 pKa = 11.84KK13 pKa = 8.88KK14 pKa = 8.64VHH16 pKa = 5.57GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.18NGRR28 pKa = 11.84KK29 pKa = 8.91VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.81GRR39 pKa = 11.84KK40 pKa = 8.75VLSAA44 pKa = 4.05
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1079243 |
38 |
2767 |
292.3 |
32.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.267 ± 0.04 | 0.812 ± 0.013 |
4.941 ± 0.029 | 7.004 ± 0.046 |
3.979 ± 0.03 | 7.753 ± 0.039 |
2.084 ± 0.021 | 6.693 ± 0.032 |
5.606 ± 0.039 | 10.109 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.753 ± 0.02 | 3.537 ± 0.022 |
4.297 ± 0.024 | 3.891 ± 0.029 |
5.481 ± 0.033 | 5.497 ± 0.03 |
5.259 ± 0.021 | 7.781 ± 0.038 |
1.172 ± 0.02 | 3.084 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
