
Glycomyces sp. MMG10089
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Glycomycetales; Glycomycetaceae; Glycomyces; unclassified Glycomyces
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
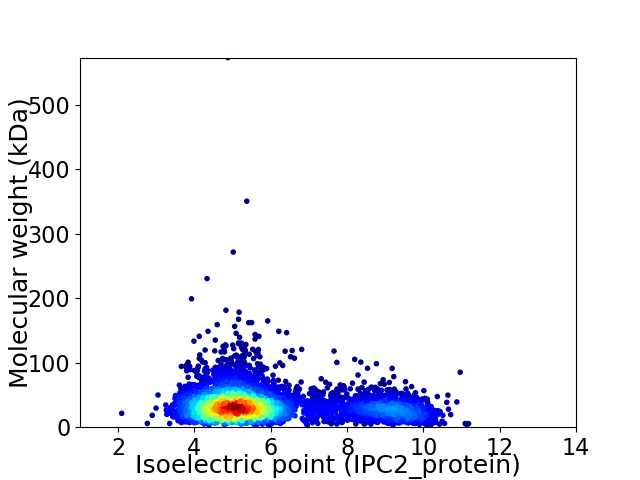
Virtual 2D-PAGE plot for 4346 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R7PWP9|A0A4R7PWP9_9ACTN MoxR-like ATPase OS=Glycomyces sp. MMG10089 OX=2135788 GN=C8R39_4135 PE=4 SV=1
MM1 pKa = 6.74QAPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84LALTGVAVGALAAGGLTFVSANQALAEE36 pKa = 4.32EE37 pKa = 5.67GCQVDD42 pKa = 4.13YY43 pKa = 11.57NVISTWGDD51 pKa = 3.12GFQANVSITAGEE63 pKa = 4.84AINGWNLQWNFPSGTTVSSAWNVDD87 pKa = 2.84WSQSGTTFTGSDD99 pKa = 3.62VGWNASIASGQTTEE113 pKa = 3.78VFGFIGSGSSTAPAQFSINGDD134 pKa = 3.56VCNGQVDD141 pKa = 4.74PDD143 pKa = 4.1PTDD146 pKa = 4.5PDD148 pKa = 3.92PTDD151 pKa = 4.88PDD153 pKa = 3.98PTDD156 pKa = 4.57PDD158 pKa = 4.71PDD160 pKa = 4.44PGNPGDD166 pKa = 4.87KK167 pKa = 10.38VDD169 pKa = 3.79NPYY172 pKa = 10.9VGAQVYY178 pKa = 8.71VNPIWSANAAAEE190 pKa = 4.31PGGSAIANEE199 pKa = 4.2PTGVWLDD206 pKa = 3.58QTSAIYY212 pKa = 10.72GNGSPTTGSYY222 pKa = 11.26GLADD226 pKa = 4.18HH227 pKa = 7.16LDD229 pKa = 3.72EE230 pKa = 5.62AVSQGADD237 pKa = 4.06LIQVVIYY244 pKa = 9.21NLPGRR249 pKa = 11.84DD250 pKa = 3.5CAALASNGQLAPNEE264 pKa = 3.72INRR267 pKa = 11.84YY268 pKa = 5.96KK269 pKa = 10.31TEE271 pKa = 4.4YY272 pKa = 9.42IDD274 pKa = 4.45EE275 pKa = 4.57IVDD278 pKa = 3.51IMSDD282 pKa = 3.2PAYY285 pKa = 10.68ADD287 pKa = 3.42LRR289 pKa = 11.84IVNVIEE295 pKa = 4.65IDD297 pKa = 3.71SLPNLITNVSPRR309 pKa = 11.84EE310 pKa = 4.09TATDD314 pKa = 3.32TCDD317 pKa = 2.98TMLANGGYY325 pKa = 9.99VDD327 pKa = 4.02GVSYY331 pKa = 10.85AAAEE335 pKa = 4.27LGALPNTYY343 pKa = 10.68NYY345 pKa = 9.95IDD347 pKa = 4.45AGHH350 pKa = 6.89HH351 pKa = 5.7GWIGWGDD358 pKa = 4.18DD359 pKa = 3.33NPEE362 pKa = 3.82YY363 pKa = 11.38DD364 pKa = 3.7NFRR367 pKa = 11.84ASAALFGSLIGQNGMTADD385 pKa = 4.11DD386 pKa = 3.54VHH388 pKa = 7.93GFISNTANYY397 pKa = 10.08SVLEE401 pKa = 4.0EE402 pKa = 4.88PYY404 pKa = 10.16WDD406 pKa = 3.13VDD408 pKa = 3.86QNVGGQALNQNPDD421 pKa = 3.97LPWVDD426 pKa = 2.72WNDD429 pKa = 3.4YY430 pKa = 11.1NNEE433 pKa = 3.89KK434 pKa = 10.92DD435 pKa = 3.33FTEE438 pKa = 4.13ALRR441 pKa = 11.84TEE443 pKa = 5.27LIANGFNSDD452 pKa = 2.87IGMLIDD458 pKa = 3.66TSRR461 pKa = 11.84NGWGGSYY468 pKa = 10.31RR469 pKa = 11.84PTGPSTSNDD478 pKa = 2.71PVTFVDD484 pKa = 4.93EE485 pKa = 4.2SRR487 pKa = 11.84IDD489 pKa = 3.47QRR491 pKa = 11.84IQKK494 pKa = 10.01GNWCNQAGAGLGEE507 pKa = 4.31RR508 pKa = 11.84PQVQPEE514 pKa = 4.0PWIDD518 pKa = 3.13AYY520 pKa = 11.47VWIKK524 pKa = 10.65PPGEE528 pKa = 4.04SDD530 pKa = 3.36GSSEE534 pKa = 6.07LIDD537 pKa = 3.69NDD539 pKa = 3.3EE540 pKa = 4.37GKK542 pKa = 10.93GFDD545 pKa = 3.89EE546 pKa = 4.9MCDD549 pKa = 3.32PTYY552 pKa = 11.1GGNPRR557 pKa = 11.84NGNNLSGARR566 pKa = 11.84GDD568 pKa = 4.01MPVSGHH574 pKa = 5.69WSSEE578 pKa = 3.76QFQEE582 pKa = 4.25LLQNAWPPINN592 pKa = 3.92
MM1 pKa = 6.74QAPRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84LALTGVAVGALAAGGLTFVSANQALAEE36 pKa = 4.32EE37 pKa = 5.67GCQVDD42 pKa = 4.13YY43 pKa = 11.57NVISTWGDD51 pKa = 3.12GFQANVSITAGEE63 pKa = 4.84AINGWNLQWNFPSGTTVSSAWNVDD87 pKa = 2.84WSQSGTTFTGSDD99 pKa = 3.62VGWNASIASGQTTEE113 pKa = 3.78VFGFIGSGSSTAPAQFSINGDD134 pKa = 3.56VCNGQVDD141 pKa = 4.74PDD143 pKa = 4.1PTDD146 pKa = 4.5PDD148 pKa = 3.92PTDD151 pKa = 4.88PDD153 pKa = 3.98PTDD156 pKa = 4.57PDD158 pKa = 4.71PDD160 pKa = 4.44PGNPGDD166 pKa = 4.87KK167 pKa = 10.38VDD169 pKa = 3.79NPYY172 pKa = 10.9VGAQVYY178 pKa = 8.71VNPIWSANAAAEE190 pKa = 4.31PGGSAIANEE199 pKa = 4.2PTGVWLDD206 pKa = 3.58QTSAIYY212 pKa = 10.72GNGSPTTGSYY222 pKa = 11.26GLADD226 pKa = 4.18HH227 pKa = 7.16LDD229 pKa = 3.72EE230 pKa = 5.62AVSQGADD237 pKa = 4.06LIQVVIYY244 pKa = 9.21NLPGRR249 pKa = 11.84DD250 pKa = 3.5CAALASNGQLAPNEE264 pKa = 3.72INRR267 pKa = 11.84YY268 pKa = 5.96KK269 pKa = 10.31TEE271 pKa = 4.4YY272 pKa = 9.42IDD274 pKa = 4.45EE275 pKa = 4.57IVDD278 pKa = 3.51IMSDD282 pKa = 3.2PAYY285 pKa = 10.68ADD287 pKa = 3.42LRR289 pKa = 11.84IVNVIEE295 pKa = 4.65IDD297 pKa = 3.71SLPNLITNVSPRR309 pKa = 11.84EE310 pKa = 4.09TATDD314 pKa = 3.32TCDD317 pKa = 2.98TMLANGGYY325 pKa = 9.99VDD327 pKa = 4.02GVSYY331 pKa = 10.85AAAEE335 pKa = 4.27LGALPNTYY343 pKa = 10.68NYY345 pKa = 9.95IDD347 pKa = 4.45AGHH350 pKa = 6.89HH351 pKa = 5.7GWIGWGDD358 pKa = 4.18DD359 pKa = 3.33NPEE362 pKa = 3.82YY363 pKa = 11.38DD364 pKa = 3.7NFRR367 pKa = 11.84ASAALFGSLIGQNGMTADD385 pKa = 4.11DD386 pKa = 3.54VHH388 pKa = 7.93GFISNTANYY397 pKa = 10.08SVLEE401 pKa = 4.0EE402 pKa = 4.88PYY404 pKa = 10.16WDD406 pKa = 3.13VDD408 pKa = 3.86QNVGGQALNQNPDD421 pKa = 3.97LPWVDD426 pKa = 2.72WNDD429 pKa = 3.4YY430 pKa = 11.1NNEE433 pKa = 3.89KK434 pKa = 10.92DD435 pKa = 3.33FTEE438 pKa = 4.13ALRR441 pKa = 11.84TEE443 pKa = 5.27LIANGFNSDD452 pKa = 2.87IGMLIDD458 pKa = 3.66TSRR461 pKa = 11.84NGWGGSYY468 pKa = 10.31RR469 pKa = 11.84PTGPSTSNDD478 pKa = 2.71PVTFVDD484 pKa = 4.93EE485 pKa = 4.2SRR487 pKa = 11.84IDD489 pKa = 3.47QRR491 pKa = 11.84IQKK494 pKa = 10.01GNWCNQAGAGLGEE507 pKa = 4.31RR508 pKa = 11.84PQVQPEE514 pKa = 4.0PWIDD518 pKa = 3.13AYY520 pKa = 11.47VWIKK524 pKa = 10.65PPGEE528 pKa = 4.04SDD530 pKa = 3.36GSSEE534 pKa = 6.07LIDD537 pKa = 3.69NDD539 pKa = 3.3EE540 pKa = 4.37GKK542 pKa = 10.93GFDD545 pKa = 3.89EE546 pKa = 4.9MCDD549 pKa = 3.32PTYY552 pKa = 11.1GGNPRR557 pKa = 11.84NGNNLSGARR566 pKa = 11.84GDD568 pKa = 4.01MPVSGHH574 pKa = 5.69WSSEE578 pKa = 3.76QFQEE582 pKa = 4.25LLQNAWPPINN592 pKa = 3.92
Molecular weight: 63.26 kDa
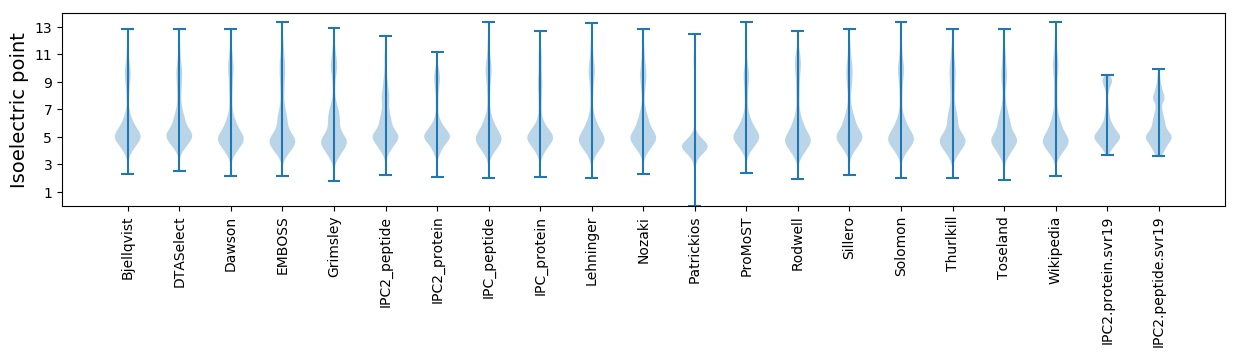
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V3F8E9|A0A4V3F8E9_9ACTN ATP-dependent Clp protease adapter protein ClpS OS=Glycomyces sp. MMG10089 OX=2135788 GN=clpS PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1448852 |
31 |
5306 |
333.4 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.809 ± 0.053 | 0.731 ± 0.011 |
6.263 ± 0.032 | 6.473 ± 0.031 |
3.096 ± 0.022 | 9.081 ± 0.039 |
2.122 ± 0.019 | 3.858 ± 0.026 |
2.136 ± 0.025 | 10.047 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.796 ± 0.015 | 2.062 ± 0.02 |
5.701 ± 0.037 | 2.683 ± 0.019 |
7.399 ± 0.043 | 5.092 ± 0.028 |
5.758 ± 0.025 | 8.084 ± 0.037 |
1.653 ± 0.019 | 2.155 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
