
Lachnoclostridium sp. An118
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; unclassified Lachnoclostridium
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
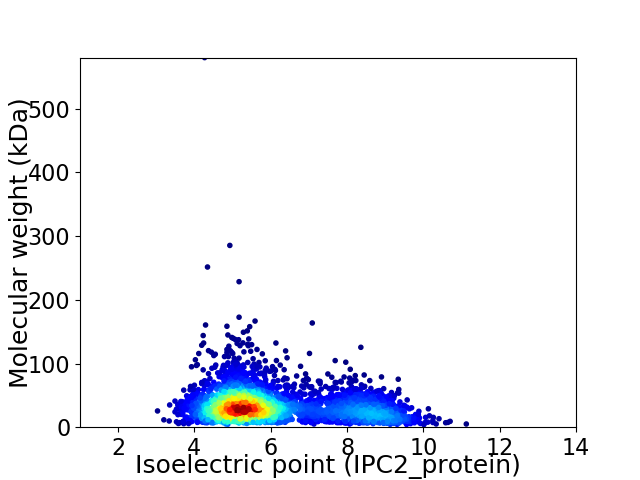
Virtual 2D-PAGE plot for 3184 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4U3U2|A0A1Y4U3U2_9FIRM 50S ribosomal protein L20 OS=Lachnoclostridium sp. An118 OX=1965547 GN=rplT PE=3 SV=1
MM1 pKa = 7.98DD2 pKa = 5.9KK3 pKa = 10.39IKK5 pKa = 10.9HH6 pKa = 5.66SFNSIASKK14 pKa = 10.12QGSYY18 pKa = 10.88SVGLTALVIAIVVLINLIVGQLPSQIRR45 pKa = 11.84QIDD48 pKa = 3.23ISTNNIYY55 pKa = 10.59EE56 pKa = 3.98ITDD59 pKa = 3.39TSRR62 pKa = 11.84EE63 pKa = 4.1LLDD66 pKa = 4.11GLDD69 pKa = 3.43RR70 pKa = 11.84DD71 pKa = 4.02VTFTVLADD79 pKa = 3.56KK80 pKa = 11.06SSTDD84 pKa = 3.26EE85 pKa = 4.74RR86 pKa = 11.84ISNFIDD92 pKa = 3.74RR93 pKa = 11.84YY94 pKa = 9.66SALSSHH100 pKa = 6.96INVEE104 pKa = 4.01WIDD107 pKa = 3.95PVLHH111 pKa = 6.6PSALTTYY118 pKa = 10.42SADD121 pKa = 2.98SDD123 pKa = 4.57TIVVSCDD130 pKa = 2.98DD131 pKa = 3.9TDD133 pKa = 4.01RR134 pKa = 11.84STTVAFSDD142 pKa = 3.73IITYY146 pKa = 10.95DD147 pKa = 3.1EE148 pKa = 3.9MSYY151 pKa = 9.28YY152 pKa = 7.97TTGSATEE159 pKa = 4.24SEE161 pKa = 4.48FDD163 pKa = 4.43GEE165 pKa = 4.66GQLSSAVNYY174 pKa = 7.29VTTDD178 pKa = 3.04VTRR181 pKa = 11.84NVYY184 pKa = 8.17YY185 pKa = 10.43TSGHH189 pKa = 6.04GEE191 pKa = 4.05SSLSSSVTDD200 pKa = 3.97LLGKK204 pKa = 10.44SSLTLTEE211 pKa = 4.87LNLMMASSIPEE222 pKa = 4.34DD223 pKa = 3.98CDD225 pKa = 4.14LLLMDD230 pKa = 5.69APANDD235 pKa = 3.71LSEE238 pKa = 6.09DD239 pKa = 3.99EE240 pKa = 5.04VSMLTDD246 pKa = 3.29YY247 pKa = 11.02MDD249 pKa = 4.14GGGHH253 pKa = 7.21LFVILGSDD261 pKa = 3.89VSQTPNLASFLEE273 pKa = 4.93TYY275 pKa = 10.83GLTSAGGYY283 pKa = 8.31IADD286 pKa = 4.13MQRR289 pKa = 11.84SYY291 pKa = 11.07QGNYY295 pKa = 9.74YY296 pKa = 10.48YY297 pKa = 10.05IFPEE301 pKa = 4.25LSVSGDD307 pKa = 3.48MANEE311 pKa = 4.05LQSDD315 pKa = 3.92MVLLVNANGLVEE327 pKa = 4.24GTPSRR332 pKa = 11.84DD333 pKa = 3.65TITLTPFMTTSSSGYY348 pKa = 10.52AVTEE352 pKa = 4.8DD353 pKa = 3.7GQSTQGTYY361 pKa = 11.1VLGATATEE369 pKa = 3.94NDD371 pKa = 3.18SRR373 pKa = 11.84LTVISSEE380 pKa = 4.23SMIDD384 pKa = 3.36EE385 pKa = 5.09TITSQFTNIEE395 pKa = 3.96NLTLFMNAVTANFDD409 pKa = 4.29DD410 pKa = 4.32IEE412 pKa = 4.31NLSIEE417 pKa = 4.39PKK419 pKa = 10.35SLQIEE424 pKa = 4.81YY425 pKa = 8.42NTMQYY430 pKa = 9.99TGLIGLAAIVGIPVVFLLYY449 pKa = 10.73GLRR452 pKa = 11.84RR453 pKa = 11.84WMKK456 pKa = 9.67RR457 pKa = 11.84RR458 pKa = 11.84KK459 pKa = 9.4AA460 pKa = 3.44
MM1 pKa = 7.98DD2 pKa = 5.9KK3 pKa = 10.39IKK5 pKa = 10.9HH6 pKa = 5.66SFNSIASKK14 pKa = 10.12QGSYY18 pKa = 10.88SVGLTALVIAIVVLINLIVGQLPSQIRR45 pKa = 11.84QIDD48 pKa = 3.23ISTNNIYY55 pKa = 10.59EE56 pKa = 3.98ITDD59 pKa = 3.39TSRR62 pKa = 11.84EE63 pKa = 4.1LLDD66 pKa = 4.11GLDD69 pKa = 3.43RR70 pKa = 11.84DD71 pKa = 4.02VTFTVLADD79 pKa = 3.56KK80 pKa = 11.06SSTDD84 pKa = 3.26EE85 pKa = 4.74RR86 pKa = 11.84ISNFIDD92 pKa = 3.74RR93 pKa = 11.84YY94 pKa = 9.66SALSSHH100 pKa = 6.96INVEE104 pKa = 4.01WIDD107 pKa = 3.95PVLHH111 pKa = 6.6PSALTTYY118 pKa = 10.42SADD121 pKa = 2.98SDD123 pKa = 4.57TIVVSCDD130 pKa = 2.98DD131 pKa = 3.9TDD133 pKa = 4.01RR134 pKa = 11.84STTVAFSDD142 pKa = 3.73IITYY146 pKa = 10.95DD147 pKa = 3.1EE148 pKa = 3.9MSYY151 pKa = 9.28YY152 pKa = 7.97TTGSATEE159 pKa = 4.24SEE161 pKa = 4.48FDD163 pKa = 4.43GEE165 pKa = 4.66GQLSSAVNYY174 pKa = 7.29VTTDD178 pKa = 3.04VTRR181 pKa = 11.84NVYY184 pKa = 8.17YY185 pKa = 10.43TSGHH189 pKa = 6.04GEE191 pKa = 4.05SSLSSSVTDD200 pKa = 3.97LLGKK204 pKa = 10.44SSLTLTEE211 pKa = 4.87LNLMMASSIPEE222 pKa = 4.34DD223 pKa = 3.98CDD225 pKa = 4.14LLLMDD230 pKa = 5.69APANDD235 pKa = 3.71LSEE238 pKa = 6.09DD239 pKa = 3.99EE240 pKa = 5.04VSMLTDD246 pKa = 3.29YY247 pKa = 11.02MDD249 pKa = 4.14GGGHH253 pKa = 7.21LFVILGSDD261 pKa = 3.89VSQTPNLASFLEE273 pKa = 4.93TYY275 pKa = 10.83GLTSAGGYY283 pKa = 8.31IADD286 pKa = 4.13MQRR289 pKa = 11.84SYY291 pKa = 11.07QGNYY295 pKa = 9.74YY296 pKa = 10.48YY297 pKa = 10.05IFPEE301 pKa = 4.25LSVSGDD307 pKa = 3.48MANEE311 pKa = 4.05LQSDD315 pKa = 3.92MVLLVNANGLVEE327 pKa = 4.24GTPSRR332 pKa = 11.84DD333 pKa = 3.65TITLTPFMTTSSSGYY348 pKa = 10.52AVTEE352 pKa = 4.8DD353 pKa = 3.7GQSTQGTYY361 pKa = 11.1VLGATATEE369 pKa = 3.94NDD371 pKa = 3.18SRR373 pKa = 11.84LTVISSEE380 pKa = 4.23SMIDD384 pKa = 3.36EE385 pKa = 5.09TITSQFTNIEE395 pKa = 3.96NLTLFMNAVTANFDD409 pKa = 4.29DD410 pKa = 4.32IEE412 pKa = 4.31NLSIEE417 pKa = 4.39PKK419 pKa = 10.35SLQIEE424 pKa = 4.81YY425 pKa = 8.42NTMQYY430 pKa = 9.99TGLIGLAAIVGIPVVFLLYY449 pKa = 10.73GLRR452 pKa = 11.84RR453 pKa = 11.84WMKK456 pKa = 9.67RR457 pKa = 11.84RR458 pKa = 11.84KK459 pKa = 9.4AA460 pKa = 3.44
Molecular weight: 50.17 kDa
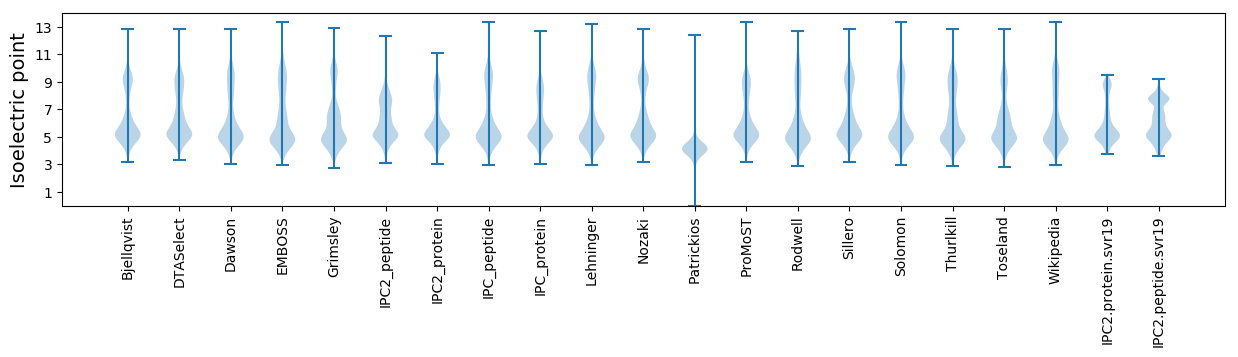
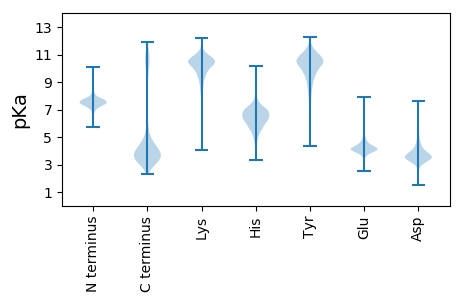
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4TVF3|A0A1Y4TVF3_9FIRM Uncharacterized protein OS=Lachnoclostridium sp. An118 OX=1965547 GN=B5E62_12990 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.79KK9 pKa = 7.6RR10 pKa = 11.84QRR12 pKa = 11.84AKK14 pKa = 9.2VHH16 pKa = 5.65GFRR19 pKa = 11.84SRR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.71VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.1GRR39 pKa = 11.84KK40 pKa = 8.83KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1015018 |
35 |
5317 |
318.8 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.929 ± 0.047 | 1.535 ± 0.018 |
5.694 ± 0.036 | 8.048 ± 0.049 |
3.965 ± 0.029 | 7.606 ± 0.041 |
1.762 ± 0.021 | 6.645 ± 0.041 |
5.855 ± 0.045 | 9.238 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.091 ± 0.025 | 3.495 ± 0.032 |
3.492 ± 0.026 | 3.331 ± 0.024 |
5.592 ± 0.047 | 5.593 ± 0.035 |
5.271 ± 0.04 | 6.955 ± 0.037 |
0.926 ± 0.015 | 3.979 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
