
Cottontail rabbit papillomavirus (strain Kansas) (CRPV) (Papillomavirus sylvilagi)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Kappapapillomavirus; Kappapapillomavirus 2
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
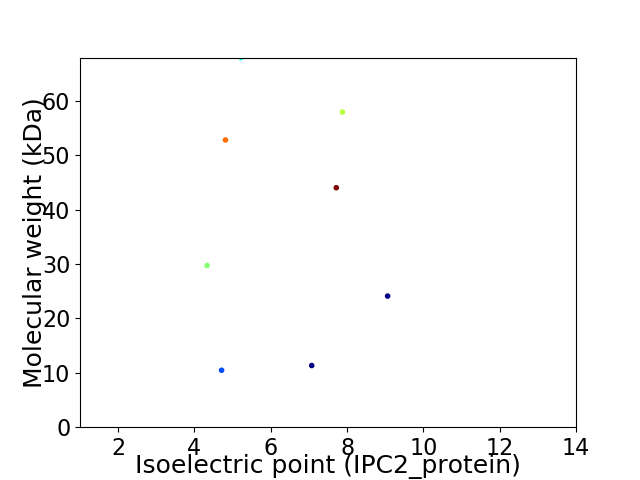
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03130|VE7_CRPVK Protein E7 OS=Cottontail rabbit papillomavirus (strain Kansas) OX=31553 GN=E7 PE=3 SV=1
MM1 pKa = 7.5EE2 pKa = 5.02NCLPRR7 pKa = 11.84SLEE10 pKa = 3.82KK11 pKa = 10.61LQQILQISLEE21 pKa = 4.34DD22 pKa = 4.02LPFGCIFCGKK32 pKa = 10.03LLGAAEE38 pKa = 4.16KK39 pKa = 10.76QLFKK43 pKa = 10.79CTGLCIVWHH52 pKa = 6.4KK53 pKa = 9.79GWPYY57 pKa = 8.87GTCRR61 pKa = 11.84DD62 pKa = 4.11CTVLSCALDD71 pKa = 4.39LYY73 pKa = 11.12CHH75 pKa = 7.3LALTAPALEE84 pKa = 4.69AEE86 pKa = 4.33ALVGQEE92 pKa = 3.3ISSWFMRR99 pKa = 11.84CTVCGRR105 pKa = 11.84RR106 pKa = 11.84LTIPEE111 pKa = 4.5KK112 pKa = 10.52IEE114 pKa = 3.5LRR116 pKa = 11.84ARR118 pKa = 11.84NCTLCCIDD126 pKa = 4.05KK127 pKa = 10.68GQYY130 pKa = 7.9FQWRR134 pKa = 11.84GHH136 pKa = 6.16CSSCKK141 pKa = 10.4LSDD144 pKa = 3.24QGDD147 pKa = 3.7LGGYY151 pKa = 8.23PPSPGSRR158 pKa = 11.84CGEE161 pKa = 3.91CDD163 pKa = 3.34EE164 pKa = 5.23CCVPDD169 pKa = 4.23LTHH172 pKa = 6.64LTPVDD177 pKa = 4.08LEE179 pKa = 4.26EE180 pKa = 5.51LGLYY184 pKa = 8.44PGPEE188 pKa = 3.75GTYY191 pKa = 10.24PDD193 pKa = 5.08LVDD196 pKa = 5.1LGPGVFGEE204 pKa = 4.16EE205 pKa = 4.24DD206 pKa = 3.63EE207 pKa = 4.64EE208 pKa = 5.83GGGLFDD214 pKa = 5.0SFEE217 pKa = 4.45EE218 pKa = 4.26EE219 pKa = 4.6DD220 pKa = 4.45PGPNQCGCFFCTSYY234 pKa = 11.19PSGTGDD240 pKa = 3.15TDD242 pKa = 3.57INQGPAGAAGIALQSDD258 pKa = 4.13PVCFCEE264 pKa = 3.77NCINFTEE271 pKa = 4.7FRR273 pKa = 3.87
MM1 pKa = 7.5EE2 pKa = 5.02NCLPRR7 pKa = 11.84SLEE10 pKa = 3.82KK11 pKa = 10.61LQQILQISLEE21 pKa = 4.34DD22 pKa = 4.02LPFGCIFCGKK32 pKa = 10.03LLGAAEE38 pKa = 4.16KK39 pKa = 10.76QLFKK43 pKa = 10.79CTGLCIVWHH52 pKa = 6.4KK53 pKa = 9.79GWPYY57 pKa = 8.87GTCRR61 pKa = 11.84DD62 pKa = 4.11CTVLSCALDD71 pKa = 4.39LYY73 pKa = 11.12CHH75 pKa = 7.3LALTAPALEE84 pKa = 4.69AEE86 pKa = 4.33ALVGQEE92 pKa = 3.3ISSWFMRR99 pKa = 11.84CTVCGRR105 pKa = 11.84RR106 pKa = 11.84LTIPEE111 pKa = 4.5KK112 pKa = 10.52IEE114 pKa = 3.5LRR116 pKa = 11.84ARR118 pKa = 11.84NCTLCCIDD126 pKa = 4.05KK127 pKa = 10.68GQYY130 pKa = 7.9FQWRR134 pKa = 11.84GHH136 pKa = 6.16CSSCKK141 pKa = 10.4LSDD144 pKa = 3.24QGDD147 pKa = 3.7LGGYY151 pKa = 8.23PPSPGSRR158 pKa = 11.84CGEE161 pKa = 3.91CDD163 pKa = 3.34EE164 pKa = 5.23CCVPDD169 pKa = 4.23LTHH172 pKa = 6.64LTPVDD177 pKa = 4.08LEE179 pKa = 4.26EE180 pKa = 5.51LGLYY184 pKa = 8.44PGPEE188 pKa = 3.75GTYY191 pKa = 10.24PDD193 pKa = 5.08LVDD196 pKa = 5.1LGPGVFGEE204 pKa = 4.16EE205 pKa = 4.24DD206 pKa = 3.63EE207 pKa = 4.64EE208 pKa = 5.83GGGLFDD214 pKa = 5.0SFEE217 pKa = 4.45EE218 pKa = 4.26EE219 pKa = 4.6DD220 pKa = 4.45PGPNQCGCFFCTSYY234 pKa = 11.19PSGTGDD240 pKa = 3.15TDD242 pKa = 3.57INQGPAGAAGIALQSDD258 pKa = 4.13PVCFCEE264 pKa = 3.77NCINFTEE271 pKa = 4.7FRR273 pKa = 3.87
Molecular weight: 29.74 kDa
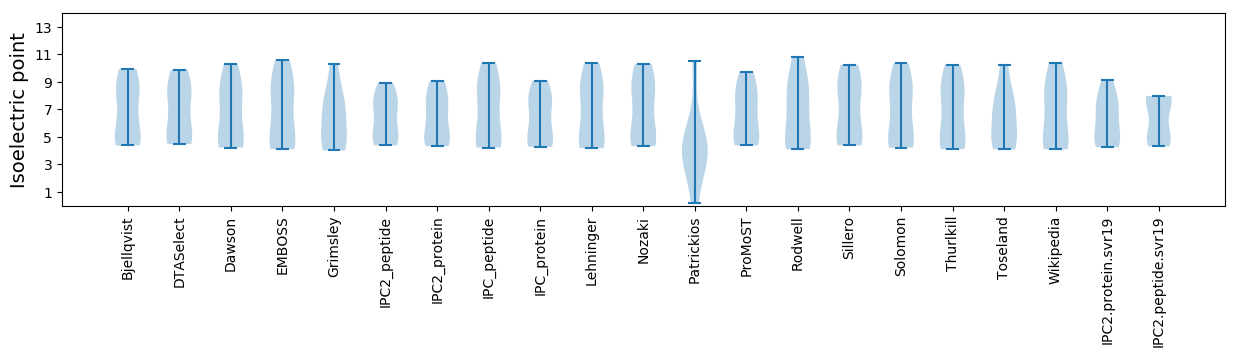
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03125|VE5_CRPVK Probable protein E5 OS=Cottontail rabbit papillomavirus (strain Kansas) OX=31553 GN=E5 PE=3 SV=1
MM1 pKa = 7.28SHH3 pKa = 5.24GHH5 pKa = 5.86CRR7 pKa = 11.84IPVEE11 pKa = 4.48KK12 pKa = 10.44GSKK15 pKa = 9.77ALRR18 pKa = 11.84KK19 pKa = 8.71RR20 pKa = 11.84HH21 pKa = 5.27SKK23 pKa = 8.81RR24 pKa = 11.84TQLLLRR30 pKa = 11.84FTMMVTEE37 pKa = 4.58GTTMNTHH44 pKa = 6.43CGVYY48 pKa = 10.48LLLGTLMGSGLRR60 pKa = 11.84LKK62 pKa = 10.74VEE64 pKa = 3.72WTIEE68 pKa = 3.95GFIMWTLKK76 pKa = 9.61EE77 pKa = 4.5TMCIMWTSQPTRR89 pKa = 11.84DD90 pKa = 3.82VLLLMDD96 pKa = 4.06TMTWCFKK103 pKa = 9.96TCASLLLSPAPPSRR117 pKa = 11.84WSVPLKK123 pKa = 9.8TPSPKK128 pKa = 10.09RR129 pKa = 11.84PPTVQCPPLKK139 pKa = 10.13RR140 pKa = 11.84KK141 pKa = 9.52QGPKK145 pKa = 9.95PRR147 pKa = 11.84VHH149 pKa = 6.75WADD152 pKa = 3.41EE153 pKa = 4.55GQGHH157 pKa = 5.49QGCNEE162 pKa = 3.48GRR164 pKa = 11.84QSNEE168 pKa = 3.15NRR170 pKa = 11.84PPRR173 pKa = 11.84TKK175 pKa = 10.47RR176 pKa = 11.84ILLPGTSDD184 pKa = 3.73RR185 pKa = 11.84LLQRR189 pKa = 11.84TLDD192 pKa = 3.55EE193 pKa = 3.94EE194 pKa = 4.18LRR196 pKa = 11.84RR197 pKa = 11.84LEE199 pKa = 3.82EE200 pKa = 4.23HH201 pKa = 6.68LPGGIDD207 pKa = 3.73GFASLL212 pKa = 4.98
MM1 pKa = 7.28SHH3 pKa = 5.24GHH5 pKa = 5.86CRR7 pKa = 11.84IPVEE11 pKa = 4.48KK12 pKa = 10.44GSKK15 pKa = 9.77ALRR18 pKa = 11.84KK19 pKa = 8.71RR20 pKa = 11.84HH21 pKa = 5.27SKK23 pKa = 8.81RR24 pKa = 11.84TQLLLRR30 pKa = 11.84FTMMVTEE37 pKa = 4.58GTTMNTHH44 pKa = 6.43CGVYY48 pKa = 10.48LLLGTLMGSGLRR60 pKa = 11.84LKK62 pKa = 10.74VEE64 pKa = 3.72WTIEE68 pKa = 3.95GFIMWTLKK76 pKa = 9.61EE77 pKa = 4.5TMCIMWTSQPTRR89 pKa = 11.84DD90 pKa = 3.82VLLLMDD96 pKa = 4.06TMTWCFKK103 pKa = 9.96TCASLLLSPAPPSRR117 pKa = 11.84WSVPLKK123 pKa = 9.8TPSPKK128 pKa = 10.09RR129 pKa = 11.84PPTVQCPPLKK139 pKa = 10.13RR140 pKa = 11.84KK141 pKa = 9.52QGPKK145 pKa = 9.95PRR147 pKa = 11.84VHH149 pKa = 6.75WADD152 pKa = 3.41EE153 pKa = 4.55GQGHH157 pKa = 5.49QGCNEE162 pKa = 3.48GRR164 pKa = 11.84QSNEE168 pKa = 3.15NRR170 pKa = 11.84PPRR173 pKa = 11.84TKK175 pKa = 10.47RR176 pKa = 11.84ILLPGTSDD184 pKa = 3.73RR185 pKa = 11.84LLQRR189 pKa = 11.84TLDD192 pKa = 3.55EE193 pKa = 3.94EE194 pKa = 4.18LRR196 pKa = 11.84RR197 pKa = 11.84LEE199 pKa = 3.82EE200 pKa = 4.23HH201 pKa = 6.68LPGGIDD207 pKa = 3.73GFASLL212 pKa = 4.98
Molecular weight: 24.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2669 |
94 |
602 |
333.6 |
37.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.032 ± 0.63 | 2.698 ± 0.938 |
6.107 ± 0.528 | 6.145 ± 0.488 |
4.159 ± 0.373 | 6.744 ± 0.787 |
2.136 ± 0.263 | 3.784 ± 0.3 |
4.946 ± 0.719 | 9.816 ± 0.869 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.873 ± 0.405 | 3.634 ± 0.372 |
6.294 ± 0.619 | 4.871 ± 0.484 |
5.807 ± 0.56 | 7.981 ± 0.625 |
6.332 ± 0.499 | 6.22 ± 0.476 |
1.349 ± 0.268 | 3.072 ± 0.507 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
