
Saccharothrix syringae (Nocardiopsis syringae)
Taxonomy: cellular organisms;
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
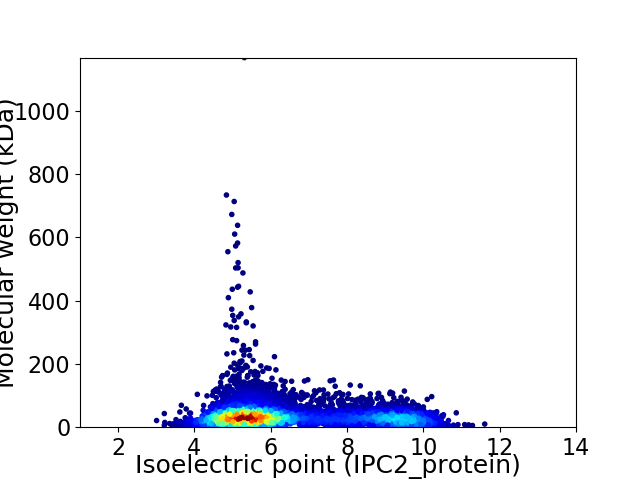
Virtual 2D-PAGE plot for 8856 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q0GZ91|A0A5Q0GZ91_SACSY GlsB/YeaQ/YmgE family stress response membrane protein OS=Saccharothrix syringae OX=103733 GN=EKG83_16605 PE=3 SV=1
MM1 pKa = 7.58PRR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 6.08RR6 pKa = 11.84NEE8 pKa = 3.89VALSTTPTSRR18 pKa = 11.84PRR20 pKa = 11.84TAVVRR25 pKa = 11.84RR26 pKa = 11.84LRR28 pKa = 11.84VLGSATTLALASLVAAGAPAHH49 pKa = 6.41AAFPGDD55 pKa = 3.06NGKK58 pKa = 9.81IYY60 pKa = 9.13FTEE63 pKa = 4.11SGNLYY68 pKa = 8.96TVNPDD73 pKa = 2.92GSGVTQVLGSPGANAPRR90 pKa = 11.84VNAQGTKK97 pKa = 9.02VAFEE101 pKa = 4.54RR102 pKa = 11.84GVNSPDD108 pKa = 3.56LQVHH112 pKa = 5.2VMNPDD117 pKa = 2.96GTGITAITSGAGADD131 pKa = 3.99GQSLAWSPDD140 pKa = 2.96GTKK143 pKa = 10.58LIYY146 pKa = 10.38RR147 pKa = 11.84PGGTLTVINADD158 pKa = 3.5GSNPTPLGVSGNHH171 pKa = 6.45PNWSPDD177 pKa = 2.65GSKK180 pKa = 9.9IVYY183 pKa = 9.43QDD185 pKa = 2.96SGVIKK190 pKa = 9.45TVNPDD195 pKa = 2.87GTGQTTLASTPGVTYY210 pKa = 9.02YY211 pKa = 11.17APNWSPDD218 pKa = 2.96SSKK221 pKa = 9.44ITFTAFSFDD230 pKa = 3.46PLGNHH235 pKa = 6.14VYY237 pKa = 10.33TMNADD242 pKa = 3.64GTNQTSLADD251 pKa = 4.24GYY253 pKa = 11.39NSVWSPDD260 pKa = 3.13GTKK263 pKa = 9.66IQYY266 pKa = 9.72QDD268 pKa = 3.42NNGLLLMNPDD278 pKa = 3.23GTNQTRR284 pKa = 11.84LYY286 pKa = 9.09TFTFGIVTTGDD297 pKa = 2.95WAAIPPVGADD307 pKa = 3.16VSVSVTDD314 pKa = 3.47SADD317 pKa = 3.59PVSLGGDD324 pKa = 3.56PYY326 pKa = 11.33SYY328 pKa = 10.08TVSVANAGPAGATGVAVSTTLSGAAASIDD357 pKa = 3.77SATSSQGSCSVSAPTVSCTLGGVAASGSATVTITVTPSATGTVTATSTVSATGTDD412 pKa = 3.63PTPGNNTDD420 pKa = 3.82AEE422 pKa = 4.57STTVNNALGCTITGTNGPDD441 pKa = 3.16TLTGTNGNDD450 pKa = 3.69VICGLGGNDD459 pKa = 4.23TISGGNGADD468 pKa = 3.62TIYY471 pKa = 11.08AGAGNDD477 pKa = 4.67AINGGNGNDD486 pKa = 3.41TVYY489 pKa = 11.06AGAGDD494 pKa = 3.84DD495 pKa = 3.56SSYY498 pKa = 11.9GEE500 pKa = 4.81DD501 pKa = 4.9LLGLLDD507 pKa = 4.41NGADD511 pKa = 3.56TLYY514 pKa = 10.86GGPGNDD520 pKa = 3.98TLDD523 pKa = 3.88GQNGNDD529 pKa = 3.44ILYY532 pKa = 10.46DD533 pKa = 3.55HH534 pKa = 7.88DD535 pKa = 4.24GTDD538 pKa = 3.51TLSGGVGNDD547 pKa = 3.8SIDD550 pKa = 3.52VQDD553 pKa = 4.18GVGGDD558 pKa = 3.68TANGGLGTDD567 pKa = 3.33TCTVDD572 pKa = 3.83GGDD575 pKa = 3.42ITTSCC580 pKa = 4.24
MM1 pKa = 7.58PRR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 6.08RR6 pKa = 11.84NEE8 pKa = 3.89VALSTTPTSRR18 pKa = 11.84PRR20 pKa = 11.84TAVVRR25 pKa = 11.84RR26 pKa = 11.84LRR28 pKa = 11.84VLGSATTLALASLVAAGAPAHH49 pKa = 6.41AAFPGDD55 pKa = 3.06NGKK58 pKa = 9.81IYY60 pKa = 9.13FTEE63 pKa = 4.11SGNLYY68 pKa = 8.96TVNPDD73 pKa = 2.92GSGVTQVLGSPGANAPRR90 pKa = 11.84VNAQGTKK97 pKa = 9.02VAFEE101 pKa = 4.54RR102 pKa = 11.84GVNSPDD108 pKa = 3.56LQVHH112 pKa = 5.2VMNPDD117 pKa = 2.96GTGITAITSGAGADD131 pKa = 3.99GQSLAWSPDD140 pKa = 2.96GTKK143 pKa = 10.58LIYY146 pKa = 10.38RR147 pKa = 11.84PGGTLTVINADD158 pKa = 3.5GSNPTPLGVSGNHH171 pKa = 6.45PNWSPDD177 pKa = 2.65GSKK180 pKa = 9.9IVYY183 pKa = 9.43QDD185 pKa = 2.96SGVIKK190 pKa = 9.45TVNPDD195 pKa = 2.87GTGQTTLASTPGVTYY210 pKa = 9.02YY211 pKa = 11.17APNWSPDD218 pKa = 2.96SSKK221 pKa = 9.44ITFTAFSFDD230 pKa = 3.46PLGNHH235 pKa = 6.14VYY237 pKa = 10.33TMNADD242 pKa = 3.64GTNQTSLADD251 pKa = 4.24GYY253 pKa = 11.39NSVWSPDD260 pKa = 3.13GTKK263 pKa = 9.66IQYY266 pKa = 9.72QDD268 pKa = 3.42NNGLLLMNPDD278 pKa = 3.23GTNQTRR284 pKa = 11.84LYY286 pKa = 9.09TFTFGIVTTGDD297 pKa = 2.95WAAIPPVGADD307 pKa = 3.16VSVSVTDD314 pKa = 3.47SADD317 pKa = 3.59PVSLGGDD324 pKa = 3.56PYY326 pKa = 11.33SYY328 pKa = 10.08TVSVANAGPAGATGVAVSTTLSGAAASIDD357 pKa = 3.77SATSSQGSCSVSAPTVSCTLGGVAASGSATVTITVTPSATGTVTATSTVSATGTDD412 pKa = 3.63PTPGNNTDD420 pKa = 3.82AEE422 pKa = 4.57STTVNNALGCTITGTNGPDD441 pKa = 3.16TLTGTNGNDD450 pKa = 3.69VICGLGGNDD459 pKa = 4.23TISGGNGADD468 pKa = 3.62TIYY471 pKa = 11.08AGAGNDD477 pKa = 4.67AINGGNGNDD486 pKa = 3.41TVYY489 pKa = 11.06AGAGDD494 pKa = 3.84DD495 pKa = 3.56SSYY498 pKa = 11.9GEE500 pKa = 4.81DD501 pKa = 4.9LLGLLDD507 pKa = 4.41NGADD511 pKa = 3.56TLYY514 pKa = 10.86GGPGNDD520 pKa = 3.98TLDD523 pKa = 3.88GQNGNDD529 pKa = 3.44ILYY532 pKa = 10.46DD533 pKa = 3.55HH534 pKa = 7.88DD535 pKa = 4.24GTDD538 pKa = 3.51TLSGGVGNDD547 pKa = 3.8SIDD550 pKa = 3.52VQDD553 pKa = 4.18GVGGDD558 pKa = 3.68TANGGLGTDD567 pKa = 3.33TCTVDD572 pKa = 3.83GGDD575 pKa = 3.42ITTSCC580 pKa = 4.24
Molecular weight: 57.67 kDa
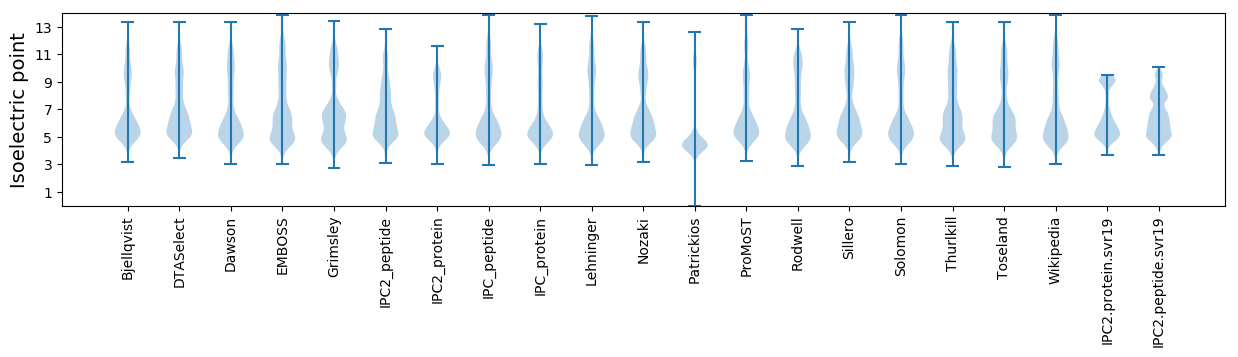
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q0HDM0|A0A5Q0HDM0_SACSY Uncharacterized protein OS=Saccharothrix syringae OX=103733 GN=EKG83_06205 PE=4 SV=1
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 8.7THH19 pKa = 5.15GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.77GRR42 pKa = 11.84KK43 pKa = 7.91QLSAA47 pKa = 3.9
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 8.7THH19 pKa = 5.15GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.77GRR42 pKa = 11.84KK43 pKa = 7.91QLSAA47 pKa = 3.9
Molecular weight: 5.48 kDa
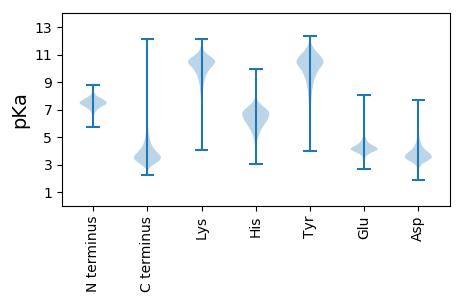
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3125302 |
24 |
11053 |
352.9 |
37.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.884 ± 0.044 | 0.783 ± 0.008 |
6.259 ± 0.021 | 5.419 ± 0.023 |
2.754 ± 0.015 | 9.527 ± 0.027 |
2.441 ± 0.014 | 2.394 ± 0.015 |
1.495 ± 0.016 | 10.608 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.386 ± 0.009 | 1.747 ± 0.017 |
6.254 ± 0.025 | 2.412 ± 0.015 |
8.848 ± 0.027 | 4.549 ± 0.02 |
5.894 ± 0.029 | 9.882 ± 0.028 |
1.585 ± 0.012 | 1.88 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
