
Chitinophaga sancti
Taxonomy: cellular organisms;
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
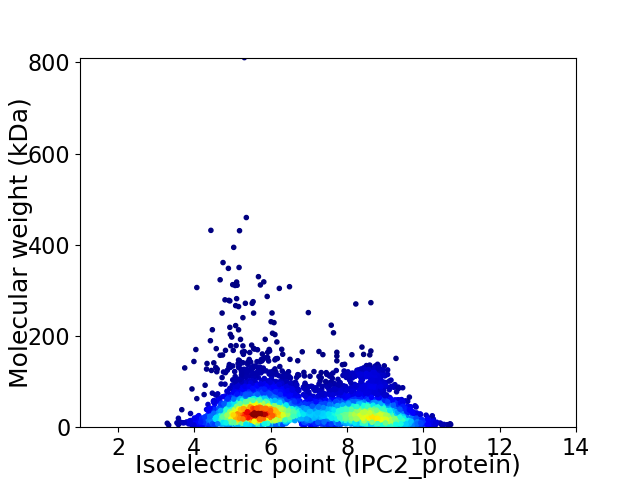
Virtual 2D-PAGE plot for 6697 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1K1SXU0|A0A1K1SXU0_9BACT Arabinose-5-phosphate isomerase OS=Chitinophaga sancti OX=1004 GN=SAMN05661012_06366 PE=3 SV=1
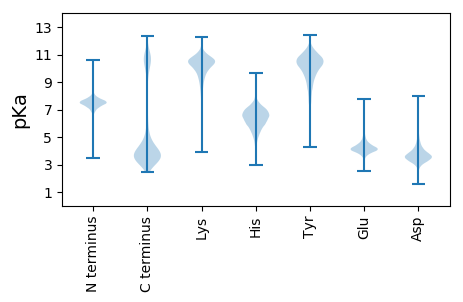
VV1 pKa = 6.14TVVEE5 pKa = 4.28DD6 pKa = 4.06TPTSFEE12 pKa = 3.9VRR14 pKa = 11.84TNDD17 pKa = 3.07TDD19 pKa = 3.93VEE21 pKa = 4.42GDD23 pKa = 3.49ALTATVIAGPVNGTVVPNPDD43 pKa = 3.11GSFKK47 pKa = 10.09YY48 pKa = 9.2TPNANYY54 pKa = 10.58NGLDD58 pKa = 3.31SLKK61 pKa = 10.97YY62 pKa = 8.46QVCDD66 pKa = 2.72NGTPSKK72 pKa = 10.67CDD74 pKa = 2.84TGTVIFTVTPVNDD87 pKa = 3.48APVAVDD93 pKa = 3.63DD94 pKa = 4.61AVTVTEE100 pKa = 4.38DD101 pKa = 3.31TPTDD105 pKa = 3.74FEE107 pKa = 4.73VRR109 pKa = 11.84TNDD112 pKa = 3.09TDD114 pKa = 3.93VEE116 pKa = 4.42GDD118 pKa = 3.49ALTATVITGPVHH130 pKa = 5.43GTVSQKK136 pKa = 10.54PDD138 pKa = 2.65GSYY141 pKa = 10.23TYY143 pKa = 10.58TPALNYY149 pKa = 10.61NGLDD153 pKa = 3.31SLKK156 pKa = 10.91YY157 pKa = 8.48QVCDD161 pKa = 2.56SGTPSKK167 pKa = 10.87CDD169 pKa = 2.85TGTVIFTVTPVNDD182 pKa = 3.59SPVAVDD188 pKa = 3.71DD189 pKa = 4.19AVTVVEE195 pKa = 4.39DD196 pKa = 4.06TPTSFEE202 pKa = 3.9VRR204 pKa = 11.84TNDD207 pKa = 3.07TDD209 pKa = 3.93VEE211 pKa = 4.42GDD213 pKa = 3.49ALTATVIAGPVNGTVVPNPDD233 pKa = 2.38GSFRR237 pKa = 11.84YY238 pKa = 7.81TPNANYY244 pKa = 10.6NGLDD248 pKa = 3.31SLKK251 pKa = 10.97YY252 pKa = 8.46QVCDD256 pKa = 2.72NGTPSKK262 pKa = 10.67CDD264 pKa = 2.87TGTVIFKK271 pKa = 7.52VTPVNDD277 pKa = 3.43APVAVDD283 pKa = 3.52DD284 pKa = 4.45AVTVVEE290 pKa = 4.59DD291 pKa = 4.03TPTDD295 pKa = 3.66FDD297 pKa = 4.15VRR299 pKa = 11.84TNDD302 pKa = 2.95TDD304 pKa = 4.02AEE306 pKa = 4.38GDD308 pKa = 3.6ALTANVISGPVHH320 pKa = 6.95GIISQNPDD328 pKa = 2.29GSFKK332 pKa = 10.07YY333 pKa = 9.2TPNANYY339 pKa = 10.58NGLDD343 pKa = 3.33SLKK346 pKa = 10.72YY347 pKa = 9.48RR348 pKa = 11.84VCDD351 pKa = 3.19NGTPSMCNTGTVIFTVTPVNDD372 pKa = 3.48APVAVDD378 pKa = 3.52DD379 pKa = 4.45AVTVVEE385 pKa = 4.59DD386 pKa = 4.03TPTDD390 pKa = 3.69FDD392 pKa = 3.93VRR394 pKa = 11.84ANDD397 pKa = 3.54TDD399 pKa = 3.95VEE401 pKa = 4.44GDD403 pKa = 3.49ALTATVITGPVNGTVSPNPDD423 pKa = 2.71GSFKK427 pKa = 10.09YY428 pKa = 9.2TPNANYY434 pKa = 10.58NGLDD438 pKa = 3.31SLKK441 pKa = 10.97YY442 pKa = 8.46QVCDD446 pKa = 2.93NGTPSMCNTGTVIFTVTPVNDD467 pKa = 3.38APIAVDD473 pKa = 3.6DD474 pKa = 4.44AVTVTEE480 pKa = 4.48DD481 pKa = 3.27TPTDD485 pKa = 3.59FDD487 pKa = 4.15VRR489 pKa = 11.84TNDD492 pKa = 3.13TDD494 pKa = 3.89VEE496 pKa = 4.42GDD498 pKa = 3.49ALTATVITGPVNGTVSQNPDD518 pKa = 2.51GSFKK522 pKa = 10.07YY523 pKa = 9.2TPNANYY529 pKa = 10.58NGLDD533 pKa = 3.31SLKK536 pKa = 10.97YY537 pKa = 8.46QVCDD541 pKa = 2.72NGTPSKK547 pKa = 10.67CDD549 pKa = 2.84TGTVIFTVTPVNDD562 pKa = 3.38APIAVDD568 pKa = 3.42DD569 pKa = 4.31AVPVVEE575 pKa = 4.64DD576 pKa = 4.03TPTSFEE582 pKa = 3.9VRR584 pKa = 11.84TNDD587 pKa = 2.87TDD589 pKa = 4.12AEE591 pKa = 4.4GDD593 pKa = 3.63ALTATVITGPVNGTVSQNPDD613 pKa = 2.51GSFKK617 pKa = 10.07YY618 pKa = 9.2TPNANYY624 pKa = 10.58NGLDD628 pKa = 3.36SLKK631 pKa = 10.65YY632 pKa = 9.42RR633 pKa = 11.84VYY635 pKa = 11.52DD636 pKa = 3.25NGTSSKK642 pKa = 10.39CDD644 pKa = 2.91TGTVIFTVTPVNDD657 pKa = 3.38APIAVDD663 pKa = 3.6DD664 pKa = 4.44AVTVTEE670 pKa = 4.26DD671 pKa = 3.38TPTSFEE677 pKa = 3.9VRR679 pKa = 11.84TNDD682 pKa = 3.07TDD684 pKa = 3.93VEE686 pKa = 4.42GDD688 pKa = 3.49ALTATVITGPVHH700 pKa = 5.73GTVSPNPDD708 pKa = 2.71GSFKK712 pKa = 10.09YY713 pKa = 9.2TPNANYY719 pKa = 10.58NGLDD723 pKa = 3.31SLKK726 pKa = 10.97YY727 pKa = 8.46QVCDD731 pKa = 2.93NGTPSMCNTGTVIFTVTPVNDD752 pKa = 3.38APIAVDD758 pKa = 3.6DD759 pKa = 4.44AVTVTEE765 pKa = 5.18DD766 pKa = 3.26APTDD770 pKa = 4.01FNVLANDD777 pKa = 3.67TDD779 pKa = 4.12VEE781 pKa = 4.42GDD783 pKa = 3.49ALTATVIISPLHH795 pKa = 4.57GTITLKK801 pKa = 11.23GNGLYY806 pKa = 9.99TYY808 pKa = 9.97TPAANYY814 pKa = 10.2NGLDD818 pKa = 3.35SLKK821 pKa = 10.91YY822 pKa = 8.48QVCDD826 pKa = 2.62SGTPGKK832 pKa = 10.33CDD834 pKa = 2.87TGTVIFTVTPVNDD847 pKa = 3.67APAGTDD853 pKa = 3.21QTITIPKK860 pKa = 7.34NTTYY864 pKa = 10.49TGTIKK869 pKa = 10.84GSDD872 pKa = 3.36ADD874 pKa = 4.28GDD876 pKa = 4.01VLTYY880 pKa = 10.69SQASSAAHH888 pKa = 6.29GNASVNPDD896 pKa = 2.52GSFTYY901 pKa = 10.2KK902 pKa = 10.7PNTDD906 pKa = 3.33YY907 pKa = 11.53VGNDD911 pKa = 3.01SFTITISDD919 pKa = 3.57GHH921 pKa = 7.23LSTTVTINITVTQVVAHH938 pKa = 6.45PGITLVKK945 pKa = 8.87TAEE948 pKa = 4.15LKK950 pKa = 11.28GNVITYY956 pKa = 7.54TFTTTNTGDD965 pKa = 3.28VALTNVILADD975 pKa = 3.54PMVNLNTTISTTLQPGEE992 pKa = 4.54VVTQTTTYY1000 pKa = 10.48TITQTDD1006 pKa = 3.91RR1007 pKa = 11.84EE1008 pKa = 4.29NGSVSNTASVSGTSSDD1024 pKa = 3.35HH1025 pKa = 5.86KK1026 pKa = 9.65TVADD1030 pKa = 3.8ISGTSANNDD1039 pKa = 2.91IPTTITVPGVIKK1051 pKa = 10.96ANDD1054 pKa = 3.62DD1055 pKa = 3.8EE1056 pKa = 5.8YY1057 pKa = 10.68DD1058 pKa = 3.35TRR1060 pKa = 11.84MNTHH1064 pKa = 6.58INFNPLNNDD1073 pKa = 3.08NLGTSTLQTINTVTTVQHH1091 pKa = 5.0GTITFDD1097 pKa = 3.26VNGNFEE1103 pKa = 4.21YY1104 pKa = 10.74TPDD1107 pKa = 3.1QGYY1110 pKa = 10.4IGDD1113 pKa = 3.89DD1114 pKa = 3.25TFTYY1118 pKa = 10.07TITDD1122 pKa = 3.24ADD1124 pKa = 4.33GYY1126 pKa = 9.74VSNIATVTIHH1136 pKa = 6.71IGAVYY1141 pKa = 10.13IKK1143 pKa = 10.43VPTLFTPNGDD1153 pKa = 3.39GKK1155 pKa = 10.63NDD1157 pKa = 3.17VFEE1160 pKa = 4.45IVGLDD1165 pKa = 3.19KK1166 pKa = 11.16YY1167 pKa = 9.46TDD1169 pKa = 3.44NEE1171 pKa = 4.45LIIVNRR1177 pKa = 11.84WGNEE1181 pKa = 3.46VYY1183 pKa = 10.22RR1184 pKa = 11.84QHH1186 pKa = 6.87NYY1188 pKa = 9.89SNTWNGDD1195 pKa = 3.27GLSEE1199 pKa = 3.86GTYY1202 pKa = 10.6YY1203 pKa = 11.33YY1204 pKa = 11.18LLRR1207 pKa = 11.84VKK1209 pKa = 10.47KK1210 pKa = 9.92QDD1212 pKa = 3.37SAEE1215 pKa = 3.93WEE1217 pKa = 4.32VFKK1220 pKa = 11.3GFTTILRR1227 pKa = 11.84KK1228 pKa = 9.75LNKK1231 pKa = 9.92
VV1 pKa = 6.14TVVEE5 pKa = 4.28DD6 pKa = 4.06TPTSFEE12 pKa = 3.9VRR14 pKa = 11.84TNDD17 pKa = 3.07TDD19 pKa = 3.93VEE21 pKa = 4.42GDD23 pKa = 3.49ALTATVIAGPVNGTVVPNPDD43 pKa = 3.11GSFKK47 pKa = 10.09YY48 pKa = 9.2TPNANYY54 pKa = 10.58NGLDD58 pKa = 3.31SLKK61 pKa = 10.97YY62 pKa = 8.46QVCDD66 pKa = 2.72NGTPSKK72 pKa = 10.67CDD74 pKa = 2.84TGTVIFTVTPVNDD87 pKa = 3.48APVAVDD93 pKa = 3.63DD94 pKa = 4.61AVTVTEE100 pKa = 4.38DD101 pKa = 3.31TPTDD105 pKa = 3.74FEE107 pKa = 4.73VRR109 pKa = 11.84TNDD112 pKa = 3.09TDD114 pKa = 3.93VEE116 pKa = 4.42GDD118 pKa = 3.49ALTATVITGPVHH130 pKa = 5.43GTVSQKK136 pKa = 10.54PDD138 pKa = 2.65GSYY141 pKa = 10.23TYY143 pKa = 10.58TPALNYY149 pKa = 10.61NGLDD153 pKa = 3.31SLKK156 pKa = 10.91YY157 pKa = 8.48QVCDD161 pKa = 2.56SGTPSKK167 pKa = 10.87CDD169 pKa = 2.85TGTVIFTVTPVNDD182 pKa = 3.59SPVAVDD188 pKa = 3.71DD189 pKa = 4.19AVTVVEE195 pKa = 4.39DD196 pKa = 4.06TPTSFEE202 pKa = 3.9VRR204 pKa = 11.84TNDD207 pKa = 3.07TDD209 pKa = 3.93VEE211 pKa = 4.42GDD213 pKa = 3.49ALTATVIAGPVNGTVVPNPDD233 pKa = 2.38GSFRR237 pKa = 11.84YY238 pKa = 7.81TPNANYY244 pKa = 10.6NGLDD248 pKa = 3.31SLKK251 pKa = 10.97YY252 pKa = 8.46QVCDD256 pKa = 2.72NGTPSKK262 pKa = 10.67CDD264 pKa = 2.87TGTVIFKK271 pKa = 7.52VTPVNDD277 pKa = 3.43APVAVDD283 pKa = 3.52DD284 pKa = 4.45AVTVVEE290 pKa = 4.59DD291 pKa = 4.03TPTDD295 pKa = 3.66FDD297 pKa = 4.15VRR299 pKa = 11.84TNDD302 pKa = 2.95TDD304 pKa = 4.02AEE306 pKa = 4.38GDD308 pKa = 3.6ALTANVISGPVHH320 pKa = 6.95GIISQNPDD328 pKa = 2.29GSFKK332 pKa = 10.07YY333 pKa = 9.2TPNANYY339 pKa = 10.58NGLDD343 pKa = 3.33SLKK346 pKa = 10.72YY347 pKa = 9.48RR348 pKa = 11.84VCDD351 pKa = 3.19NGTPSMCNTGTVIFTVTPVNDD372 pKa = 3.48APVAVDD378 pKa = 3.52DD379 pKa = 4.45AVTVVEE385 pKa = 4.59DD386 pKa = 4.03TPTDD390 pKa = 3.69FDD392 pKa = 3.93VRR394 pKa = 11.84ANDD397 pKa = 3.54TDD399 pKa = 3.95VEE401 pKa = 4.44GDD403 pKa = 3.49ALTATVITGPVNGTVSPNPDD423 pKa = 2.71GSFKK427 pKa = 10.09YY428 pKa = 9.2TPNANYY434 pKa = 10.58NGLDD438 pKa = 3.31SLKK441 pKa = 10.97YY442 pKa = 8.46QVCDD446 pKa = 2.93NGTPSMCNTGTVIFTVTPVNDD467 pKa = 3.38APIAVDD473 pKa = 3.6DD474 pKa = 4.44AVTVTEE480 pKa = 4.48DD481 pKa = 3.27TPTDD485 pKa = 3.59FDD487 pKa = 4.15VRR489 pKa = 11.84TNDD492 pKa = 3.13TDD494 pKa = 3.89VEE496 pKa = 4.42GDD498 pKa = 3.49ALTATVITGPVNGTVSQNPDD518 pKa = 2.51GSFKK522 pKa = 10.07YY523 pKa = 9.2TPNANYY529 pKa = 10.58NGLDD533 pKa = 3.31SLKK536 pKa = 10.97YY537 pKa = 8.46QVCDD541 pKa = 2.72NGTPSKK547 pKa = 10.67CDD549 pKa = 2.84TGTVIFTVTPVNDD562 pKa = 3.38APIAVDD568 pKa = 3.42DD569 pKa = 4.31AVPVVEE575 pKa = 4.64DD576 pKa = 4.03TPTSFEE582 pKa = 3.9VRR584 pKa = 11.84TNDD587 pKa = 2.87TDD589 pKa = 4.12AEE591 pKa = 4.4GDD593 pKa = 3.63ALTATVITGPVNGTVSQNPDD613 pKa = 2.51GSFKK617 pKa = 10.07YY618 pKa = 9.2TPNANYY624 pKa = 10.58NGLDD628 pKa = 3.36SLKK631 pKa = 10.65YY632 pKa = 9.42RR633 pKa = 11.84VYY635 pKa = 11.52DD636 pKa = 3.25NGTSSKK642 pKa = 10.39CDD644 pKa = 2.91TGTVIFTVTPVNDD657 pKa = 3.38APIAVDD663 pKa = 3.6DD664 pKa = 4.44AVTVTEE670 pKa = 4.26DD671 pKa = 3.38TPTSFEE677 pKa = 3.9VRR679 pKa = 11.84TNDD682 pKa = 3.07TDD684 pKa = 3.93VEE686 pKa = 4.42GDD688 pKa = 3.49ALTATVITGPVHH700 pKa = 5.73GTVSPNPDD708 pKa = 2.71GSFKK712 pKa = 10.09YY713 pKa = 9.2TPNANYY719 pKa = 10.58NGLDD723 pKa = 3.31SLKK726 pKa = 10.97YY727 pKa = 8.46QVCDD731 pKa = 2.93NGTPSMCNTGTVIFTVTPVNDD752 pKa = 3.38APIAVDD758 pKa = 3.6DD759 pKa = 4.44AVTVTEE765 pKa = 5.18DD766 pKa = 3.26APTDD770 pKa = 4.01FNVLANDD777 pKa = 3.67TDD779 pKa = 4.12VEE781 pKa = 4.42GDD783 pKa = 3.49ALTATVIISPLHH795 pKa = 4.57GTITLKK801 pKa = 11.23GNGLYY806 pKa = 9.99TYY808 pKa = 9.97TPAANYY814 pKa = 10.2NGLDD818 pKa = 3.35SLKK821 pKa = 10.91YY822 pKa = 8.48QVCDD826 pKa = 2.62SGTPGKK832 pKa = 10.33CDD834 pKa = 2.87TGTVIFTVTPVNDD847 pKa = 3.67APAGTDD853 pKa = 3.21QTITIPKK860 pKa = 7.34NTTYY864 pKa = 10.49TGTIKK869 pKa = 10.84GSDD872 pKa = 3.36ADD874 pKa = 4.28GDD876 pKa = 4.01VLTYY880 pKa = 10.69SQASSAAHH888 pKa = 6.29GNASVNPDD896 pKa = 2.52GSFTYY901 pKa = 10.2KK902 pKa = 10.7PNTDD906 pKa = 3.33YY907 pKa = 11.53VGNDD911 pKa = 3.01SFTITISDD919 pKa = 3.57GHH921 pKa = 7.23LSTTVTINITVTQVVAHH938 pKa = 6.45PGITLVKK945 pKa = 8.87TAEE948 pKa = 4.15LKK950 pKa = 11.28GNVITYY956 pKa = 7.54TFTTTNTGDD965 pKa = 3.28VALTNVILADD975 pKa = 3.54PMVNLNTTISTTLQPGEE992 pKa = 4.54VVTQTTTYY1000 pKa = 10.48TITQTDD1006 pKa = 3.91RR1007 pKa = 11.84EE1008 pKa = 4.29NGSVSNTASVSGTSSDD1024 pKa = 3.35HH1025 pKa = 5.86KK1026 pKa = 9.65TVADD1030 pKa = 3.8ISGTSANNDD1039 pKa = 2.91IPTTITVPGVIKK1051 pKa = 10.96ANDD1054 pKa = 3.62DD1055 pKa = 3.8EE1056 pKa = 5.8YY1057 pKa = 10.68DD1058 pKa = 3.35TRR1060 pKa = 11.84MNTHH1064 pKa = 6.58INFNPLNNDD1073 pKa = 3.08NLGTSTLQTINTVTTVQHH1091 pKa = 5.0GTITFDD1097 pKa = 3.26VNGNFEE1103 pKa = 4.21YY1104 pKa = 10.74TPDD1107 pKa = 3.1QGYY1110 pKa = 10.4IGDD1113 pKa = 3.89DD1114 pKa = 3.25TFTYY1118 pKa = 10.07TITDD1122 pKa = 3.24ADD1124 pKa = 4.33GYY1126 pKa = 9.74VSNIATVTIHH1136 pKa = 6.71IGAVYY1141 pKa = 10.13IKK1143 pKa = 10.43VPTLFTPNGDD1153 pKa = 3.39GKK1155 pKa = 10.63NDD1157 pKa = 3.17VFEE1160 pKa = 4.45IVGLDD1165 pKa = 3.19KK1166 pKa = 11.16YY1167 pKa = 9.46TDD1169 pKa = 3.44NEE1171 pKa = 4.45LIIVNRR1177 pKa = 11.84WGNEE1181 pKa = 3.46VYY1183 pKa = 10.22RR1184 pKa = 11.84QHH1186 pKa = 6.87NYY1188 pKa = 9.89SNTWNGDD1195 pKa = 3.27GLSEE1199 pKa = 3.86GTYY1202 pKa = 10.6YY1203 pKa = 11.33YY1204 pKa = 11.18LLRR1207 pKa = 11.84VKK1209 pKa = 10.47KK1210 pKa = 9.92QDD1212 pKa = 3.37SAEE1215 pKa = 3.93WEE1217 pKa = 4.32VFKK1220 pKa = 11.3GFTTILRR1227 pKa = 11.84KK1228 pKa = 9.75LNKK1231 pKa = 9.92
Molecular weight: 129.98 kDa
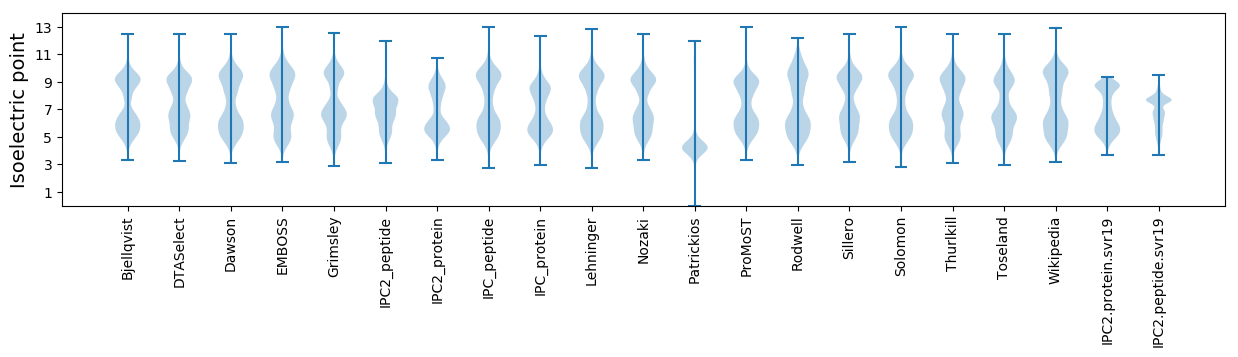
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1K1R6W4|A0A1K1R6W4_9BACT CubicO group peptidase beta-lactamase class C family OS=Chitinophaga sancti OX=1004 GN=SAMN05661012_03425 PE=4 SV=1
MM1 pKa = 7.26LRR3 pKa = 11.84WTVTFLIIALVAALFGFGGIAASAAGIAKK32 pKa = 9.97IIFFIFLVLLVISLVMGRR50 pKa = 11.84RR51 pKa = 11.84SRR53 pKa = 11.84II54 pKa = 3.41
MM1 pKa = 7.26LRR3 pKa = 11.84WTVTFLIIALVAALFGFGGIAASAAGIAKK32 pKa = 9.97IIFFIFLVLLVISLVMGRR50 pKa = 11.84RR51 pKa = 11.84SRR53 pKa = 11.84II54 pKa = 3.41
Molecular weight: 5.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2424268 |
39 |
7223 |
362.0 |
40.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.914 ± 0.034 | 0.858 ± 0.011 |
5.285 ± 0.02 | 5.363 ± 0.031 |
4.583 ± 0.02 | 6.956 ± 0.029 |
2.043 ± 0.015 | 6.776 ± 0.028 |
6.229 ± 0.029 | 9.459 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.447 ± 0.014 | 5.344 ± 0.029 |
4.046 ± 0.02 | 3.955 ± 0.018 |
4.179 ± 0.023 | 6.253 ± 0.028 |
6.16 ± 0.04 | 6.491 ± 0.024 |
1.283 ± 0.011 | 4.377 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
