
Brevundimonas viscosa
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Brevundimonas
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
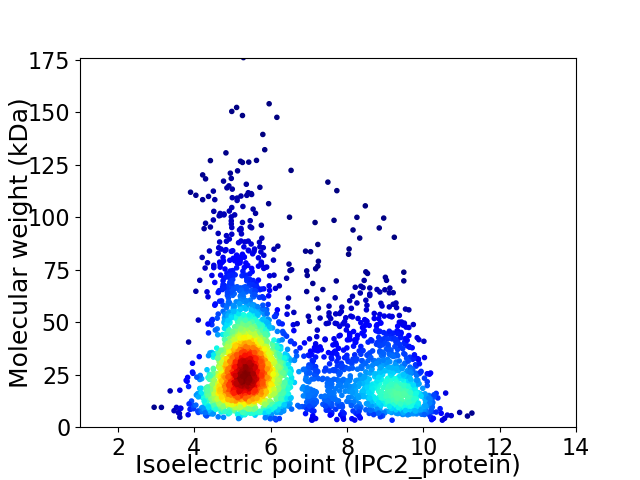
Virtual 2D-PAGE plot for 3072 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
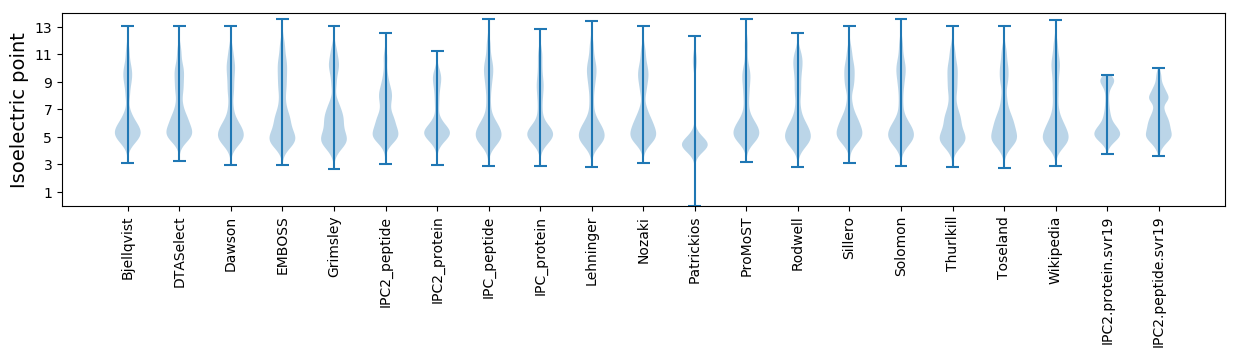
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6NRL4|A0A1I6NRL4_9CAUL Dihydrolipoyl dehydrogenase OS=Brevundimonas viscosa OX=871741 GN=SAMN05192570_0398 PE=3 SV=1
MM1 pKa = 7.55KK2 pKa = 10.28KK3 pKa = 10.52LLIAALAATLASATGAAAQDD23 pKa = 3.59ASATATYY30 pKa = 9.75GQTRR34 pKa = 11.84LRR36 pKa = 11.84SGFTPDD42 pKa = 3.1PHH44 pKa = 7.36SVRR47 pKa = 11.84ITAGGSIDD55 pKa = 3.35ASRR58 pKa = 11.84IGSPCTGSIANAPDD72 pKa = 3.39YY73 pKa = 10.84EE74 pKa = 4.45VTYY77 pKa = 9.39TAGSLPLYY85 pKa = 10.05IYY87 pKa = 10.65AQADD91 pKa = 3.41SDD93 pKa = 3.92TTLVVNGPDD102 pKa = 3.92GQWYY106 pKa = 10.43CDD108 pKa = 3.77DD109 pKa = 5.02DD110 pKa = 5.67SNGGLNPQVTFNSPRR125 pKa = 11.84SGTYY129 pKa = 9.87DD130 pKa = 2.47IWVGTFGGGTASATLHH146 pKa = 5.99ISEE149 pKa = 5.59LSDD152 pKa = 4.44DD153 pKa = 4.92DD154 pKa = 5.3SGPDD158 pKa = 3.83PDD160 pKa = 5.66DD161 pKa = 4.29SSAMPDD167 pKa = 2.89ASLRR171 pKa = 11.84ATYY174 pKa = 11.02GEE176 pKa = 3.95VSLRR180 pKa = 11.84NGFTPDD186 pKa = 3.14PHH188 pKa = 6.85RR189 pKa = 11.84VSLTAGGSVPASNVSSSCSGSIAVAPDD216 pKa = 3.27YY217 pKa = 10.94QVTYY221 pKa = 9.27TAGSLPLVFRR231 pKa = 11.84SQSSTDD237 pKa = 3.07TTLVINGPDD246 pKa = 4.13GQWSCDD252 pKa = 3.55DD253 pKa = 6.12DD254 pKa = 4.28GWQSNNAEE262 pKa = 3.97IRR264 pKa = 11.84FDD266 pKa = 3.52KK267 pKa = 10.73PMSGVYY273 pKa = 9.76DD274 pKa = 3.28VWVGVYY280 pKa = 10.59GGGTARR286 pKa = 11.84ADD288 pKa = 3.69LLVTEE293 pKa = 4.98VPP295 pKa = 3.84
MM1 pKa = 7.55KK2 pKa = 10.28KK3 pKa = 10.52LLIAALAATLASATGAAAQDD23 pKa = 3.59ASATATYY30 pKa = 9.75GQTRR34 pKa = 11.84LRR36 pKa = 11.84SGFTPDD42 pKa = 3.1PHH44 pKa = 7.36SVRR47 pKa = 11.84ITAGGSIDD55 pKa = 3.35ASRR58 pKa = 11.84IGSPCTGSIANAPDD72 pKa = 3.39YY73 pKa = 10.84EE74 pKa = 4.45VTYY77 pKa = 9.39TAGSLPLYY85 pKa = 10.05IYY87 pKa = 10.65AQADD91 pKa = 3.41SDD93 pKa = 3.92TTLVVNGPDD102 pKa = 3.92GQWYY106 pKa = 10.43CDD108 pKa = 3.77DD109 pKa = 5.02DD110 pKa = 5.67SNGGLNPQVTFNSPRR125 pKa = 11.84SGTYY129 pKa = 9.87DD130 pKa = 2.47IWVGTFGGGTASATLHH146 pKa = 5.99ISEE149 pKa = 5.59LSDD152 pKa = 4.44DD153 pKa = 4.92DD154 pKa = 5.3SGPDD158 pKa = 3.83PDD160 pKa = 5.66DD161 pKa = 4.29SSAMPDD167 pKa = 2.89ASLRR171 pKa = 11.84ATYY174 pKa = 11.02GEE176 pKa = 3.95VSLRR180 pKa = 11.84NGFTPDD186 pKa = 3.14PHH188 pKa = 6.85RR189 pKa = 11.84VSLTAGGSVPASNVSSSCSGSIAVAPDD216 pKa = 3.27YY217 pKa = 10.94QVTYY221 pKa = 9.27TAGSLPLVFRR231 pKa = 11.84SQSSTDD237 pKa = 3.07TTLVINGPDD246 pKa = 4.13GQWSCDD252 pKa = 3.55DD253 pKa = 6.12DD254 pKa = 4.28GWQSNNAEE262 pKa = 3.97IRR264 pKa = 11.84FDD266 pKa = 3.52KK267 pKa = 10.73PMSGVYY273 pKa = 9.76DD274 pKa = 3.28VWVGVYY280 pKa = 10.59GGGTARR286 pKa = 11.84ADD288 pKa = 3.69LLVTEE293 pKa = 4.98VPP295 pKa = 3.84
Molecular weight: 30.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6SML5|A0A1I6SML5_9CAUL Uncharacterized protein OS=Brevundimonas viscosa OX=871741 GN=SAMN05192570_2587 PE=4 SV=1
MM1 pKa = 8.09RR2 pKa = 11.84LPPIATVLGGLLLSRR17 pKa = 11.84GGRR20 pKa = 11.84RR21 pKa = 11.84MAGRR25 pKa = 11.84HH26 pKa = 5.07AGKK29 pKa = 9.93LALATLALQLWQSRR43 pKa = 11.84RR44 pKa = 11.84TGGAAGANRR53 pKa = 11.84PGRR56 pKa = 11.84RR57 pKa = 11.84WSGRR61 pKa = 11.84RR62 pKa = 11.84PP63 pKa = 3.26
MM1 pKa = 8.09RR2 pKa = 11.84LPPIATVLGGLLLSRR17 pKa = 11.84GGRR20 pKa = 11.84RR21 pKa = 11.84MAGRR25 pKa = 11.84HH26 pKa = 5.07AGKK29 pKa = 9.93LALATLALQLWQSRR43 pKa = 11.84RR44 pKa = 11.84TGGAAGANRR53 pKa = 11.84PGRR56 pKa = 11.84RR57 pKa = 11.84WSGRR61 pKa = 11.84RR62 pKa = 11.84PP63 pKa = 3.26
Molecular weight: 6.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
904085 |
29 |
1621 |
294.3 |
31.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.408 ± 0.074 | 0.731 ± 0.014 |
5.906 ± 0.038 | 6.04 ± 0.041 |
3.39 ± 0.031 | 9.149 ± 0.038 |
1.768 ± 0.019 | 3.988 ± 0.032 |
2.363 ± 0.034 | 10.013 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.228 ± 0.02 | 2.079 ± 0.028 |
5.791 ± 0.037 | 2.93 ± 0.022 |
8.213 ± 0.052 | 4.732 ± 0.029 |
5.025 ± 0.029 | 7.763 ± 0.038 |
1.534 ± 0.023 | 1.95 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
