
Spiribacter sp. C176
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Spiribacter; unclassified Spiribacter
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
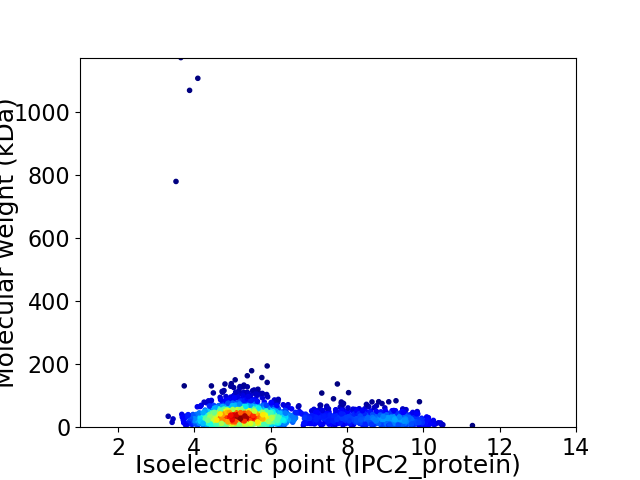
Virtual 2D-PAGE plot for 2078 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N7QTI3|A0A6N7QTI3_9GAMM NADP(H)-dependent aldo-keto reductase OS=Spiribacter sp. C176 OX=2664894 GN=GH984_09280 PE=4 SV=1
MM1 pKa = 7.34KK2 pKa = 10.18KK3 pKa = 10.45VYY5 pKa = 9.87LAAAATSALALAATPVMADD24 pKa = 3.29DD25 pKa = 4.25HH26 pKa = 8.71ASMSVYY32 pKa = 10.45GWITAEE38 pKa = 4.02VQDD41 pKa = 5.91NGTEE45 pKa = 3.77WDD47 pKa = 3.31ATGNGNSPSRR57 pKa = 11.84FGIRR61 pKa = 11.84GSRR64 pKa = 11.84EE65 pKa = 3.46FGAVTGDD72 pKa = 3.14AQLEE76 pKa = 4.36YY77 pKa = 10.8GVTKK81 pKa = 10.67AQGEE85 pKa = 4.71GPGIRR90 pKa = 11.84LANAALSGDD99 pKa = 3.79FGSVRR104 pKa = 11.84IGSQWNPGYY113 pKa = 10.85LWTTATTDD121 pKa = 3.15VFTSGAYY128 pKa = 8.73TGARR132 pKa = 11.84STDD135 pKa = 3.61FVFRR139 pKa = 11.84QDD141 pKa = 3.15GAIFYY146 pKa = 7.1YY147 pKa = 10.63APSLGGLDD155 pKa = 3.13IAVGGNMLASDD166 pKa = 4.46DD167 pKa = 4.45SSDD170 pKa = 3.86EE171 pKa = 4.0FDD173 pKa = 3.5TMTVSARR180 pKa = 11.84YY181 pKa = 9.82SFGDD185 pKa = 3.79LYY187 pKa = 11.75VSATYY192 pKa = 9.85MEE194 pKa = 5.42ADD196 pKa = 4.28DD197 pKa = 4.85EE198 pKa = 4.5NSSEE202 pKa = 4.06ATAFAASYY210 pKa = 10.49DD211 pKa = 3.57FGMFTLAGSMSDD223 pKa = 3.18NDD225 pKa = 4.13GVGSYY230 pKa = 10.8NDD232 pKa = 3.22NGTPYY237 pKa = 10.21EE238 pKa = 4.32VVATVSATEE247 pKa = 3.89EE248 pKa = 4.19LTLKK252 pKa = 10.55AAYY255 pKa = 8.46TDD257 pKa = 3.83RR258 pKa = 11.84DD259 pKa = 3.86LEE261 pKa = 5.59AGDD264 pKa = 4.47DD265 pKa = 3.54SSMAAEE271 pKa = 3.83AAYY274 pKa = 10.45AFGSGVTGFAGFSSSDD290 pKa = 3.29DD291 pKa = 4.23GINDD295 pKa = 3.85GEE297 pKa = 5.1DD298 pKa = 2.93ILSFGVQVVFF308 pKa = 4.98
MM1 pKa = 7.34KK2 pKa = 10.18KK3 pKa = 10.45VYY5 pKa = 9.87LAAAATSALALAATPVMADD24 pKa = 3.29DD25 pKa = 4.25HH26 pKa = 8.71ASMSVYY32 pKa = 10.45GWITAEE38 pKa = 4.02VQDD41 pKa = 5.91NGTEE45 pKa = 3.77WDD47 pKa = 3.31ATGNGNSPSRR57 pKa = 11.84FGIRR61 pKa = 11.84GSRR64 pKa = 11.84EE65 pKa = 3.46FGAVTGDD72 pKa = 3.14AQLEE76 pKa = 4.36YY77 pKa = 10.8GVTKK81 pKa = 10.67AQGEE85 pKa = 4.71GPGIRR90 pKa = 11.84LANAALSGDD99 pKa = 3.79FGSVRR104 pKa = 11.84IGSQWNPGYY113 pKa = 10.85LWTTATTDD121 pKa = 3.15VFTSGAYY128 pKa = 8.73TGARR132 pKa = 11.84STDD135 pKa = 3.61FVFRR139 pKa = 11.84QDD141 pKa = 3.15GAIFYY146 pKa = 7.1YY147 pKa = 10.63APSLGGLDD155 pKa = 3.13IAVGGNMLASDD166 pKa = 4.46DD167 pKa = 4.45SSDD170 pKa = 3.86EE171 pKa = 4.0FDD173 pKa = 3.5TMTVSARR180 pKa = 11.84YY181 pKa = 9.82SFGDD185 pKa = 3.79LYY187 pKa = 11.75VSATYY192 pKa = 9.85MEE194 pKa = 5.42ADD196 pKa = 4.28DD197 pKa = 4.85EE198 pKa = 4.5NSSEE202 pKa = 4.06ATAFAASYY210 pKa = 10.49DD211 pKa = 3.57FGMFTLAGSMSDD223 pKa = 3.18NDD225 pKa = 4.13GVGSYY230 pKa = 10.8NDD232 pKa = 3.22NGTPYY237 pKa = 10.21EE238 pKa = 4.32VVATVSATEE247 pKa = 3.89EE248 pKa = 4.19LTLKK252 pKa = 10.55AAYY255 pKa = 8.46TDD257 pKa = 3.83RR258 pKa = 11.84DD259 pKa = 3.86LEE261 pKa = 5.59AGDD264 pKa = 4.47DD265 pKa = 3.54SSMAAEE271 pKa = 3.83AAYY274 pKa = 10.45AFGSGVTGFAGFSSSDD290 pKa = 3.29DD291 pKa = 4.23GINDD295 pKa = 3.85GEE297 pKa = 5.1DD298 pKa = 2.93ILSFGVQVVFF308 pKa = 4.98
Molecular weight: 32.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N7QPK6|A0A6N7QPK6_9GAMM UPF0260 protein GH984_01090 OS=Spiribacter sp. C176 OX=2664894 GN=GH984_01090 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.13RR14 pKa = 11.84THH16 pKa = 6.01GFRR19 pKa = 11.84SRR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.79ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.38GRR39 pKa = 11.84ARR41 pKa = 11.84LAPP44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.52RR12 pKa = 11.84KK13 pKa = 8.13RR14 pKa = 11.84THH16 pKa = 6.01GFRR19 pKa = 11.84SRR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.79ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.38GRR39 pKa = 11.84ARR41 pKa = 11.84LAPP44 pKa = 4.03
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
708737 |
26 |
11782 |
341.1 |
37.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.525 ± 0.073 | 0.856 ± 0.031 |
5.732 ± 0.094 | 6.106 ± 0.065 |
3.451 ± 0.043 | 8.314 ± 0.176 |
2.192 ± 0.048 | 5.447 ± 0.039 |
2.65 ± 0.053 | 10.795 ± 0.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.282 ± 0.053 | 3.039 ± 0.077 |
4.788 ± 0.093 | 4.228 ± 0.048 |
6.658 ± 0.12 | 5.541 ± 0.095 |
5.377 ± 0.148 | 7.313 ± 0.052 |
1.363 ± 0.03 | 2.344 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
