
Acidobacteria bacterium AB60
Taxonomy: cellular organisms; Bacteria; Acidobacteria; unclassified Acidobacteria
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
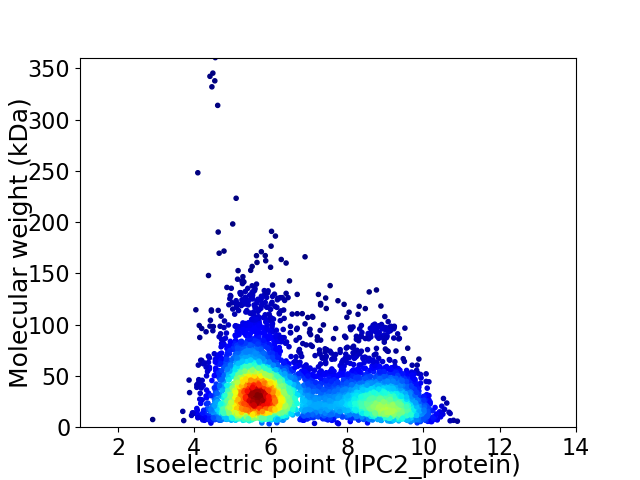
Virtual 2D-PAGE plot for 5367 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5M8SK51|A0A5M8SK51_9BACT Peroxiredoxin OS=Acidobacteria bacterium AB60 OX=1230846 GN=DYQ86_15425 PE=3 SV=1
MM1 pKa = 7.75ALGVLNNLSAIYY13 pKa = 10.65AEE15 pKa = 4.24NNLNNTSSSLQAVLRR30 pKa = 11.84QLSSGSRR37 pKa = 11.84INSGADD43 pKa = 3.18DD44 pKa = 4.03AAGLSLVNGLKK55 pKa = 10.79ANSTALAQSGTNATEE70 pKa = 4.2GVGLLQVADD79 pKa = 4.12GALSQVTSLLNRR91 pKa = 11.84AITLATEE98 pKa = 4.67ASNGTLNSSQDD109 pKa = 3.05AAADD113 pKa = 3.54QEE115 pKa = 4.61YY116 pKa = 10.75QSILAEE122 pKa = 4.13INNIGSTTTYY132 pKa = 9.01NQQRR136 pKa = 11.84VFGGGPVAIYY146 pKa = 9.86TGDD149 pKa = 3.56SSVTGASVDD158 pKa = 3.71LLNIRR163 pKa = 11.84GLSSQSVGDD172 pKa = 3.77SAGTMSYY179 pKa = 7.49TTGTNNVFIDD189 pKa = 4.23LSTAGKK195 pKa = 9.6YY196 pKa = 9.92AQTTDD201 pKa = 3.03SLAAGGSTSLEE212 pKa = 3.8VAYY215 pKa = 10.4LVAGTGTVANTTITVSGANNTVQGLIQAINNANLGITASFGTAAMAGAAAVAAVGGAANQSEE277 pKa = 4.51TGIIIAGNVNAGNAPTQAGYY297 pKa = 10.25HH298 pKa = 5.52VGIFQDD304 pKa = 3.4ATAAATDD311 pKa = 4.04LLYY314 pKa = 10.86DD315 pKa = 3.76SSVGQTFNLGVAPAASTAPQTSFTQDD341 pKa = 2.8LNAASIATISYY352 pKa = 8.61TDD354 pKa = 3.36AAGVSLSGTDD364 pKa = 3.74LKK366 pKa = 11.4SSTSAQAALSLLNQAISAVAAQDD389 pKa = 4.01GYY391 pKa = 10.84IGAQINTLNSISQVMSTQQEE411 pKa = 4.47NVVSAQNAIQATDD424 pKa = 3.54YY425 pKa = 11.45AAATSNMSKK434 pKa = 10.86FEE436 pKa = 4.18ILSQTGIAALAQANQVEE453 pKa = 4.37QEE455 pKa = 4.35VIKK458 pKa = 10.76LLQQ461 pKa = 3.54
MM1 pKa = 7.75ALGVLNNLSAIYY13 pKa = 10.65AEE15 pKa = 4.24NNLNNTSSSLQAVLRR30 pKa = 11.84QLSSGSRR37 pKa = 11.84INSGADD43 pKa = 3.18DD44 pKa = 4.03AAGLSLVNGLKK55 pKa = 10.79ANSTALAQSGTNATEE70 pKa = 4.2GVGLLQVADD79 pKa = 4.12GALSQVTSLLNRR91 pKa = 11.84AITLATEE98 pKa = 4.67ASNGTLNSSQDD109 pKa = 3.05AAADD113 pKa = 3.54QEE115 pKa = 4.61YY116 pKa = 10.75QSILAEE122 pKa = 4.13INNIGSTTTYY132 pKa = 9.01NQQRR136 pKa = 11.84VFGGGPVAIYY146 pKa = 9.86TGDD149 pKa = 3.56SSVTGASVDD158 pKa = 3.71LLNIRR163 pKa = 11.84GLSSQSVGDD172 pKa = 3.77SAGTMSYY179 pKa = 7.49TTGTNNVFIDD189 pKa = 4.23LSTAGKK195 pKa = 9.6YY196 pKa = 9.92AQTTDD201 pKa = 3.03SLAAGGSTSLEE212 pKa = 3.8VAYY215 pKa = 10.4LVAGTGTVANTTITVSGANNTVQGLIQAINNANLGITASFGTAAMAGAAAVAAVGGAANQSEE277 pKa = 4.51TGIIIAGNVNAGNAPTQAGYY297 pKa = 10.25HH298 pKa = 5.52VGIFQDD304 pKa = 3.4ATAAATDD311 pKa = 4.04LLYY314 pKa = 10.86DD315 pKa = 3.76SSVGQTFNLGVAPAASTAPQTSFTQDD341 pKa = 2.8LNAASIATISYY352 pKa = 8.61TDD354 pKa = 3.36AAGVSLSGTDD364 pKa = 3.74LKK366 pKa = 11.4SSTSAQAALSLLNQAISAVAAQDD389 pKa = 4.01GYY391 pKa = 10.84IGAQINTLNSISQVMSTQQEE411 pKa = 4.47NVVSAQNAIQATDD424 pKa = 3.54YY425 pKa = 11.45AAATSNMSKK434 pKa = 10.86FEE436 pKa = 4.18ILSQTGIAALAQANQVEE453 pKa = 4.37QEE455 pKa = 4.35VIKK458 pKa = 10.76LLQQ461 pKa = 3.54
Molecular weight: 45.97 kDa
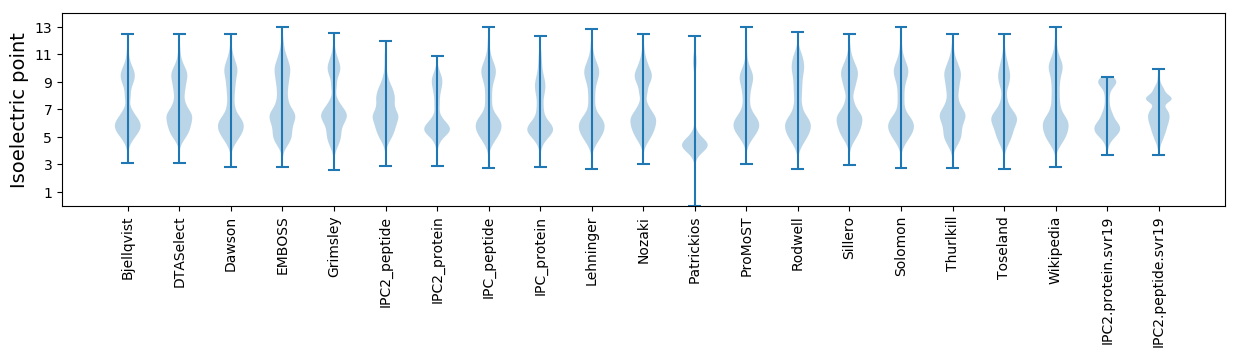
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5M8SR00|A0A5M8SR00_9BACT DUF5110 domain-containing protein OS=Acidobacteria bacterium AB60 OX=1230846 GN=DYQ86_10940 PE=3 SV=1
MM1 pKa = 7.56EE2 pKa = 5.24ARR4 pKa = 11.84FEE6 pKa = 3.96QFIRR10 pKa = 11.84EE11 pKa = 3.89RR12 pKa = 11.84QYY14 pKa = 11.32LQNVTPRR21 pKa = 11.84TIEE24 pKa = 3.58WYY26 pKa = 10.04RR27 pKa = 11.84FSLRR31 pKa = 11.84WLPCEE36 pKa = 4.39DD37 pKa = 3.55PTRR40 pKa = 11.84AQLDD44 pKa = 3.7EE45 pKa = 4.22MVVRR49 pKa = 11.84MRR51 pKa = 11.84EE52 pKa = 3.86RR53 pKa = 11.84GLKK56 pKa = 8.38PTGCNCLCRR65 pKa = 11.84AINAYY70 pKa = 9.88LKK72 pKa = 10.32WSGSTHH78 pKa = 7.29KK79 pKa = 10.15IRR81 pKa = 11.84PFKK84 pKa = 10.9EE85 pKa = 3.94PDD87 pKa = 3.15SVLPIFTEE95 pKa = 4.11QQFKK99 pKa = 10.88RR100 pKa = 11.84LIAWKK105 pKa = 9.46PKK107 pKa = 9.94SKK109 pKa = 9.93YY110 pKa = 7.05QRR112 pKa = 11.84RR113 pKa = 11.84LHH115 pKa = 6.46LLILFLLDD123 pKa = 3.22TGCRR127 pKa = 11.84ISEE130 pKa = 4.03ALGLRR135 pKa = 11.84VSEE138 pKa = 4.29IDD140 pKa = 4.61LDD142 pKa = 4.01NLLVKK147 pKa = 10.64LDD149 pKa = 3.66GKK151 pKa = 10.23GRR153 pKa = 11.84KK154 pKa = 7.56QRR156 pKa = 11.84IVPISLEE163 pKa = 3.74LRR165 pKa = 11.84KK166 pKa = 10.18AIVRR170 pKa = 11.84FCTDD174 pKa = 3.11RR175 pKa = 11.84ARR177 pKa = 11.84KK178 pKa = 8.94PDD180 pKa = 3.7LLLFASRR187 pKa = 11.84VEE189 pKa = 4.47TTWVRR194 pKa = 11.84RR195 pKa = 11.84NILRR199 pKa = 11.84DD200 pKa = 3.39VKK202 pKa = 10.45RR203 pKa = 11.84VCRR206 pKa = 11.84TLGFEE211 pKa = 3.98PPIRR215 pKa = 11.84TLHH218 pKa = 6.23AFRR221 pKa = 11.84HH222 pKa = 5.02TFAVNYY228 pKa = 9.02LRR230 pKa = 11.84RR231 pKa = 11.84GGSVFHH237 pKa = 5.81LQKK240 pKa = 10.33MLGHH244 pKa = 6.1STLEE248 pKa = 3.68MTRR251 pKa = 11.84RR252 pKa = 11.84YY253 pKa = 10.59ANLVTADD260 pKa = 3.77LQAVHH265 pKa = 6.37EE266 pKa = 4.41RR267 pKa = 11.84LSLLSRR273 pKa = 4.14
MM1 pKa = 7.56EE2 pKa = 5.24ARR4 pKa = 11.84FEE6 pKa = 3.96QFIRR10 pKa = 11.84EE11 pKa = 3.89RR12 pKa = 11.84QYY14 pKa = 11.32LQNVTPRR21 pKa = 11.84TIEE24 pKa = 3.58WYY26 pKa = 10.04RR27 pKa = 11.84FSLRR31 pKa = 11.84WLPCEE36 pKa = 4.39DD37 pKa = 3.55PTRR40 pKa = 11.84AQLDD44 pKa = 3.7EE45 pKa = 4.22MVVRR49 pKa = 11.84MRR51 pKa = 11.84EE52 pKa = 3.86RR53 pKa = 11.84GLKK56 pKa = 8.38PTGCNCLCRR65 pKa = 11.84AINAYY70 pKa = 9.88LKK72 pKa = 10.32WSGSTHH78 pKa = 7.29KK79 pKa = 10.15IRR81 pKa = 11.84PFKK84 pKa = 10.9EE85 pKa = 3.94PDD87 pKa = 3.15SVLPIFTEE95 pKa = 4.11QQFKK99 pKa = 10.88RR100 pKa = 11.84LIAWKK105 pKa = 9.46PKK107 pKa = 9.94SKK109 pKa = 9.93YY110 pKa = 7.05QRR112 pKa = 11.84RR113 pKa = 11.84LHH115 pKa = 6.46LLILFLLDD123 pKa = 3.22TGCRR127 pKa = 11.84ISEE130 pKa = 4.03ALGLRR135 pKa = 11.84VSEE138 pKa = 4.29IDD140 pKa = 4.61LDD142 pKa = 4.01NLLVKK147 pKa = 10.64LDD149 pKa = 3.66GKK151 pKa = 10.23GRR153 pKa = 11.84KK154 pKa = 7.56QRR156 pKa = 11.84IVPISLEE163 pKa = 3.74LRR165 pKa = 11.84KK166 pKa = 10.18AIVRR170 pKa = 11.84FCTDD174 pKa = 3.11RR175 pKa = 11.84ARR177 pKa = 11.84KK178 pKa = 8.94PDD180 pKa = 3.7LLLFASRR187 pKa = 11.84VEE189 pKa = 4.47TTWVRR194 pKa = 11.84RR195 pKa = 11.84NILRR199 pKa = 11.84DD200 pKa = 3.39VKK202 pKa = 10.45RR203 pKa = 11.84VCRR206 pKa = 11.84TLGFEE211 pKa = 3.98PPIRR215 pKa = 11.84TLHH218 pKa = 6.23AFRR221 pKa = 11.84HH222 pKa = 5.02TFAVNYY228 pKa = 9.02LRR230 pKa = 11.84RR231 pKa = 11.84GGSVFHH237 pKa = 5.81LQKK240 pKa = 10.33MLGHH244 pKa = 6.1STLEE248 pKa = 3.68MTRR251 pKa = 11.84RR252 pKa = 11.84YY253 pKa = 10.59ANLVTADD260 pKa = 3.77LQAVHH265 pKa = 6.37EE266 pKa = 4.41RR267 pKa = 11.84LSLLSRR273 pKa = 4.14
Molecular weight: 32.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1893627 |
29 |
3493 |
352.8 |
38.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.343 ± 0.043 | 0.923 ± 0.013 |
5.043 ± 0.023 | 5.461 ± 0.04 |
3.844 ± 0.021 | 8.24 ± 0.036 |
2.274 ± 0.017 | 4.755 ± 0.021 |
3.316 ± 0.028 | 10.014 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.013 | 3.271 ± 0.031 |
5.607 ± 0.026 | 3.827 ± 0.022 |
6.623 ± 0.039 | 6.226 ± 0.032 |
5.745 ± 0.044 | 7.187 ± 0.027 |
1.48 ± 0.016 | 2.636 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
