
Ornithinimicrobium sp. AMA3305
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ornithinimicrobiaceae; Ornithinimicrobium; unclassified Ornithinimicrobium
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
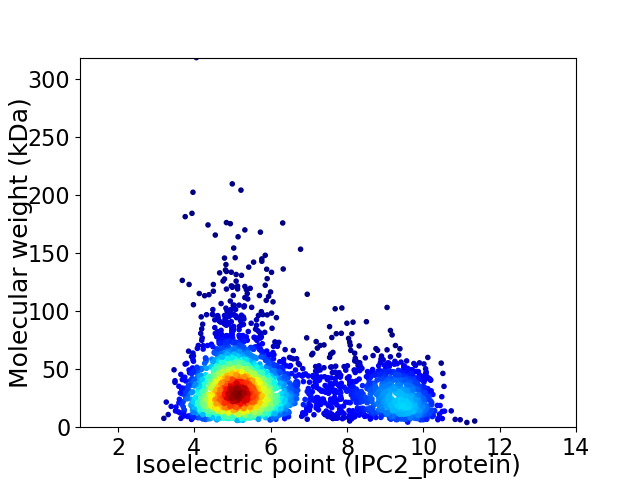
Virtual 2D-PAGE plot for 3428 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345NQA4|A0A345NQA4_9MICO Glutamyl-tRNA reductase OS=Ornithinimicrobium sp. AMA3305 OX=2283195 GN=hemA PE=3 SV=1
MM1 pKa = 7.64GGRR4 pKa = 11.84HH5 pKa = 5.77ARR7 pKa = 11.84TPRR10 pKa = 11.84LLASLLTLTLLAGCTFAGGDD30 pKa = 3.71GGAPTPTGTATQDD43 pKa = 3.05GFSTDD48 pKa = 3.53GPVTSTSTPDD58 pKa = 3.49ADD60 pKa = 3.73GAPAGLEE67 pKa = 3.96DD68 pKa = 5.84FYY70 pKa = 11.59GQQLEE75 pKa = 4.44WTDD78 pKa = 3.79CEE80 pKa = 5.12SLFEE84 pKa = 4.24CANLEE89 pKa = 4.26VPVDD93 pKa = 3.88YY94 pKa = 10.32EE95 pKa = 4.49DD96 pKa = 4.13PGGPTIEE103 pKa = 4.37LAVLRR108 pKa = 11.84AGASGEE114 pKa = 4.14PQGSLVVNPGGPGASGVDD132 pKa = 3.6YY133 pKa = 11.61AMMAGAVVSRR143 pKa = 11.84EE144 pKa = 3.4VRR146 pKa = 11.84AAYY149 pKa = 9.95DD150 pKa = 3.35VVGFDD155 pKa = 3.51PRR157 pKa = 11.84GVARR161 pKa = 11.84SAPITCFTDD170 pKa = 3.21EE171 pKa = 5.03QMDD174 pKa = 4.1TFLGTDD180 pKa = 4.47PSPDD184 pKa = 3.41DD185 pKa = 3.84PAEE188 pKa = 3.94EE189 pKa = 3.79RR190 pKa = 11.84AVEE193 pKa = 4.36GEE195 pKa = 4.36LKK197 pKa = 10.82DD198 pKa = 3.71FAQACEE204 pKa = 4.21TNAGDD209 pKa = 4.51LLGHH213 pKa = 6.23VSTVEE218 pKa = 3.53AAKK221 pKa = 11.19DD222 pKa = 3.53MDD224 pKa = 4.16VLRR227 pKa = 11.84AALGEE232 pKa = 3.93PRR234 pKa = 11.84LTYY237 pKa = 10.74LGASYY242 pKa = 8.37GTFLGTTYY250 pKa = 10.29ATLFPDD256 pKa = 3.18RR257 pKa = 11.84VGRR260 pKa = 11.84LVLDD264 pKa = 4.16GAIDD268 pKa = 3.63PTLTGLEE275 pKa = 4.12MGLGQAAGFEE285 pKa = 4.47RR286 pKa = 11.84ATRR289 pKa = 11.84AYY291 pKa = 10.72VEE293 pKa = 4.19NCVGGGSCPLGDD305 pKa = 4.8DD306 pKa = 3.44VDD308 pKa = 4.09TAMVAIPAFLDD319 pKa = 3.59EE320 pKa = 4.74VDD322 pKa = 3.68AHH324 pKa = 6.55PLPVSGDD331 pKa = 3.42TVTEE335 pKa = 4.15LTEE338 pKa = 3.88GWAMLGIIVAMYY350 pKa = 10.26SQDD353 pKa = 2.49IWPILTQALGRR364 pKa = 11.84AQQGDD369 pKa = 3.83GTLLMFLANSYY380 pKa = 11.07ASRR383 pKa = 11.84HH384 pKa = 5.28SDD386 pKa = 2.72GTYY389 pKa = 9.75TGNGMQAIYY398 pKa = 10.47AVNCLDD404 pKa = 3.73RR405 pKa = 11.84PADD408 pKa = 3.93EE409 pKa = 6.21GPDD412 pKa = 3.5LDD414 pKa = 3.94SAAVEE419 pKa = 4.31EE420 pKa = 4.37QFEE423 pKa = 4.4AASPTWGRR431 pKa = 11.84YY432 pKa = 7.51LAGDD436 pKa = 4.56GACEE440 pKa = 3.91YY441 pKa = 9.96WPVHH445 pKa = 6.52AEE447 pKa = 3.8QTLQDD452 pKa = 3.94YY453 pKa = 9.64SAQGAAPIVVIGTTRR468 pKa = 11.84DD469 pKa = 3.08PATPYY474 pKa = 10.19EE475 pKa = 4.15WAEE478 pKa = 3.9ALADD482 pKa = 3.72ILDD485 pKa = 4.04SGVLISYY492 pKa = 10.54DD493 pKa = 3.73GDD495 pKa = 3.21GHH497 pKa = 5.16TAYY500 pKa = 10.16GRR502 pKa = 11.84SNDD505 pKa = 3.98CVDD508 pKa = 4.3DD509 pKa = 4.84AVDD512 pKa = 3.93AYY514 pKa = 10.93LLEE517 pKa = 4.55GTVPQDD523 pKa = 3.9GLRR526 pKa = 11.84CC527 pKa = 3.91
MM1 pKa = 7.64GGRR4 pKa = 11.84HH5 pKa = 5.77ARR7 pKa = 11.84TPRR10 pKa = 11.84LLASLLTLTLLAGCTFAGGDD30 pKa = 3.71GGAPTPTGTATQDD43 pKa = 3.05GFSTDD48 pKa = 3.53GPVTSTSTPDD58 pKa = 3.49ADD60 pKa = 3.73GAPAGLEE67 pKa = 3.96DD68 pKa = 5.84FYY70 pKa = 11.59GQQLEE75 pKa = 4.44WTDD78 pKa = 3.79CEE80 pKa = 5.12SLFEE84 pKa = 4.24CANLEE89 pKa = 4.26VPVDD93 pKa = 3.88YY94 pKa = 10.32EE95 pKa = 4.49DD96 pKa = 4.13PGGPTIEE103 pKa = 4.37LAVLRR108 pKa = 11.84AGASGEE114 pKa = 4.14PQGSLVVNPGGPGASGVDD132 pKa = 3.6YY133 pKa = 11.61AMMAGAVVSRR143 pKa = 11.84EE144 pKa = 3.4VRR146 pKa = 11.84AAYY149 pKa = 9.95DD150 pKa = 3.35VVGFDD155 pKa = 3.51PRR157 pKa = 11.84GVARR161 pKa = 11.84SAPITCFTDD170 pKa = 3.21EE171 pKa = 5.03QMDD174 pKa = 4.1TFLGTDD180 pKa = 4.47PSPDD184 pKa = 3.41DD185 pKa = 3.84PAEE188 pKa = 3.94EE189 pKa = 3.79RR190 pKa = 11.84AVEE193 pKa = 4.36GEE195 pKa = 4.36LKK197 pKa = 10.82DD198 pKa = 3.71FAQACEE204 pKa = 4.21TNAGDD209 pKa = 4.51LLGHH213 pKa = 6.23VSTVEE218 pKa = 3.53AAKK221 pKa = 11.19DD222 pKa = 3.53MDD224 pKa = 4.16VLRR227 pKa = 11.84AALGEE232 pKa = 3.93PRR234 pKa = 11.84LTYY237 pKa = 10.74LGASYY242 pKa = 8.37GTFLGTTYY250 pKa = 10.29ATLFPDD256 pKa = 3.18RR257 pKa = 11.84VGRR260 pKa = 11.84LVLDD264 pKa = 4.16GAIDD268 pKa = 3.63PTLTGLEE275 pKa = 4.12MGLGQAAGFEE285 pKa = 4.47RR286 pKa = 11.84ATRR289 pKa = 11.84AYY291 pKa = 10.72VEE293 pKa = 4.19NCVGGGSCPLGDD305 pKa = 4.8DD306 pKa = 3.44VDD308 pKa = 4.09TAMVAIPAFLDD319 pKa = 3.59EE320 pKa = 4.74VDD322 pKa = 3.68AHH324 pKa = 6.55PLPVSGDD331 pKa = 3.42TVTEE335 pKa = 4.15LTEE338 pKa = 3.88GWAMLGIIVAMYY350 pKa = 10.26SQDD353 pKa = 2.49IWPILTQALGRR364 pKa = 11.84AQQGDD369 pKa = 3.83GTLLMFLANSYY380 pKa = 11.07ASRR383 pKa = 11.84HH384 pKa = 5.28SDD386 pKa = 2.72GTYY389 pKa = 9.75TGNGMQAIYY398 pKa = 10.47AVNCLDD404 pKa = 3.73RR405 pKa = 11.84PADD408 pKa = 3.93EE409 pKa = 6.21GPDD412 pKa = 3.5LDD414 pKa = 3.94SAAVEE419 pKa = 4.31EE420 pKa = 4.37QFEE423 pKa = 4.4AASPTWGRR431 pKa = 11.84YY432 pKa = 7.51LAGDD436 pKa = 4.56GACEE440 pKa = 3.91YY441 pKa = 9.96WPVHH445 pKa = 6.52AEE447 pKa = 3.8QTLQDD452 pKa = 3.94YY453 pKa = 9.64SAQGAAPIVVIGTTRR468 pKa = 11.84DD469 pKa = 3.08PATPYY474 pKa = 10.19EE475 pKa = 4.15WAEE478 pKa = 3.9ALADD482 pKa = 3.72ILDD485 pKa = 4.04SGVLISYY492 pKa = 10.54DD493 pKa = 3.73GDD495 pKa = 3.21GHH497 pKa = 5.16TAYY500 pKa = 10.16GRR502 pKa = 11.84SNDD505 pKa = 3.98CVDD508 pKa = 4.3DD509 pKa = 4.84AVDD512 pKa = 3.93AYY514 pKa = 10.93LLEE517 pKa = 4.55GTVPQDD523 pKa = 3.9GLRR526 pKa = 11.84CC527 pKa = 3.91
Molecular weight: 54.98 kDa
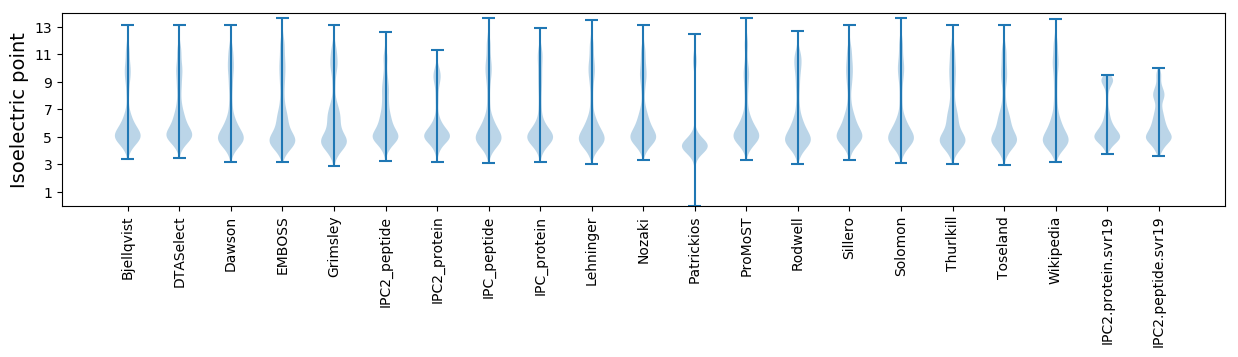
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345NP23|A0A345NP23_9MICO DUF721 domain-containing protein OS=Ornithinimicrobium sp. AMA3305 OX=2283195 GN=DV701_12205 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.65HH17 pKa = 4.47GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SALSAA45 pKa = 3.8
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84ARR15 pKa = 11.84KK16 pKa = 8.65HH17 pKa = 4.47GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SALSAA45 pKa = 3.8
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1146953 |
32 |
3005 |
334.6 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.048 ± 0.053 | 0.655 ± 0.01 |
6.256 ± 0.041 | 5.922 ± 0.038 |
2.472 ± 0.026 | 9.636 ± 0.035 |
2.31 ± 0.021 | 2.861 ± 0.035 |
1.497 ± 0.026 | 10.656 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.867 ± 0.019 | 1.412 ± 0.021 |
5.823 ± 0.033 | 3.048 ± 0.022 |
8.12 ± 0.052 | 5.0 ± 0.026 |
6.044 ± 0.029 | 9.924 ± 0.043 |
1.605 ± 0.019 | 1.843 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
